If you ever played tug-of-war in elementary school, you might remember that it isn’t the friendliest game. People fall over, hands get burned from holding on to the rope, and knees get scraped from falling on the ground. Although victory can be sweet, the injuries that come with it may make you never want to play the game again. Perhaps surprisingly, there is a similar ‘’tug-of-war” happening inside your body, as individual cells move around from one place to another in a process called cell migration. What’s more, this microscopic tug-of-war may help to heal those scrapes and bruises that happened in elementary school, and those that happen in your everyday life.
The Origin of Random Forces Inside Cells
Place yourself in a bumper car at a carnival waiting to bump into your friends. Soon enough you hear the small engine of your bumper car start and you begin to move around, bumping into anyone in your way. While the motion of your car is mostly controlled by the steering wheel, random events—like fluctuations in the motor power, your car hitting small bumps on the floor, and other cars hitting you—can affect the motion as well. What if I told you that a cell and its parts function in a similar way? Just as your car is powered by electricity, molecular motors—bio-molecules that can convert chemical energy into mechanical work—power the movement of living organisms by generating forces. In order to produce these forces, molecular motors depend on an organic molecule called ATP.
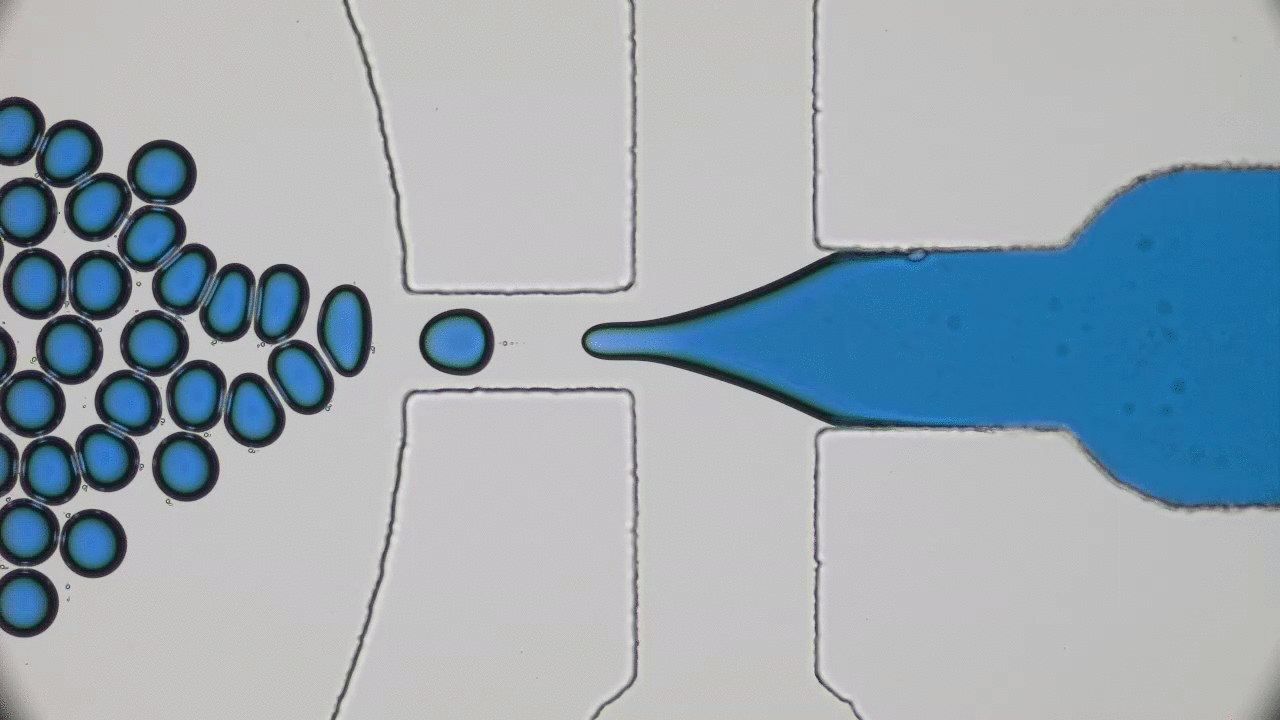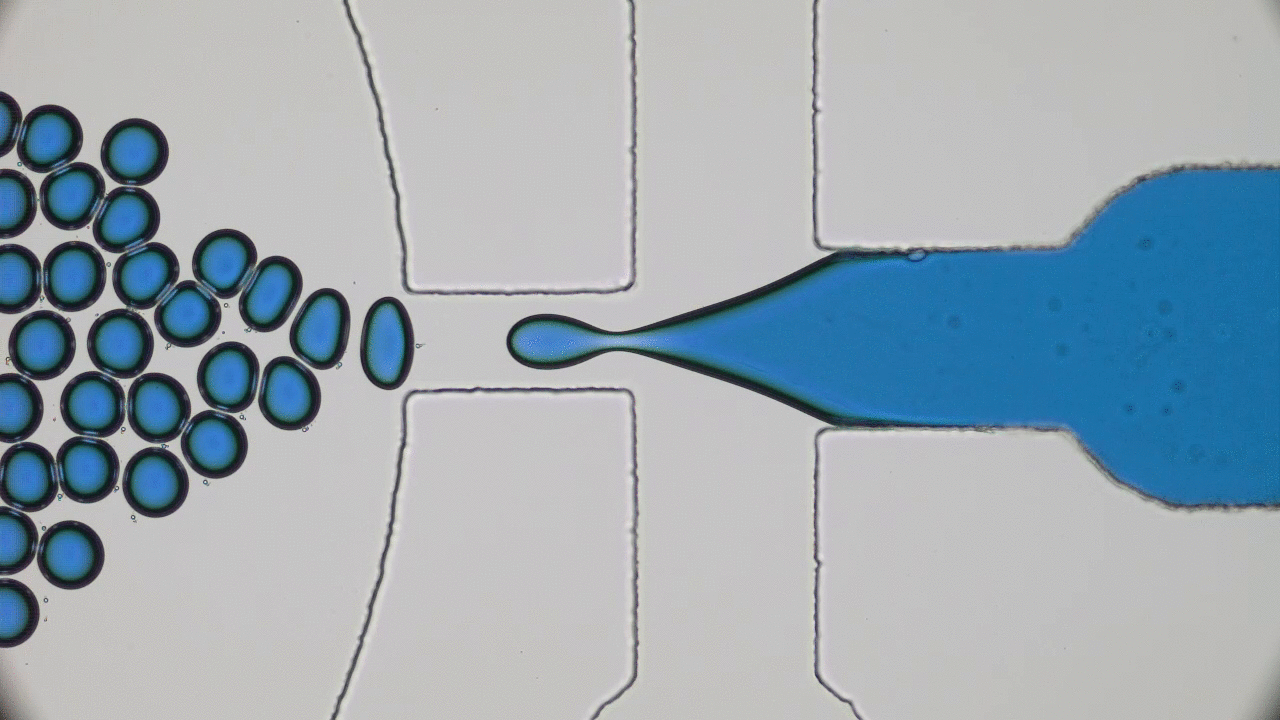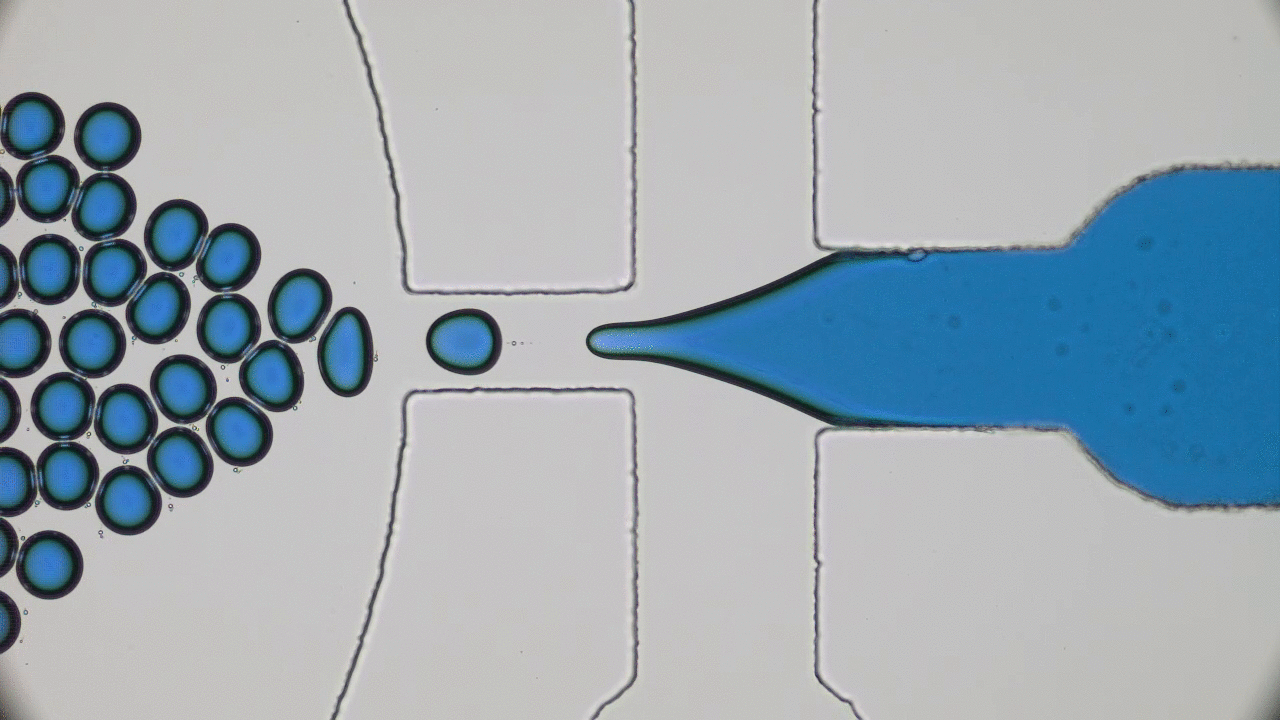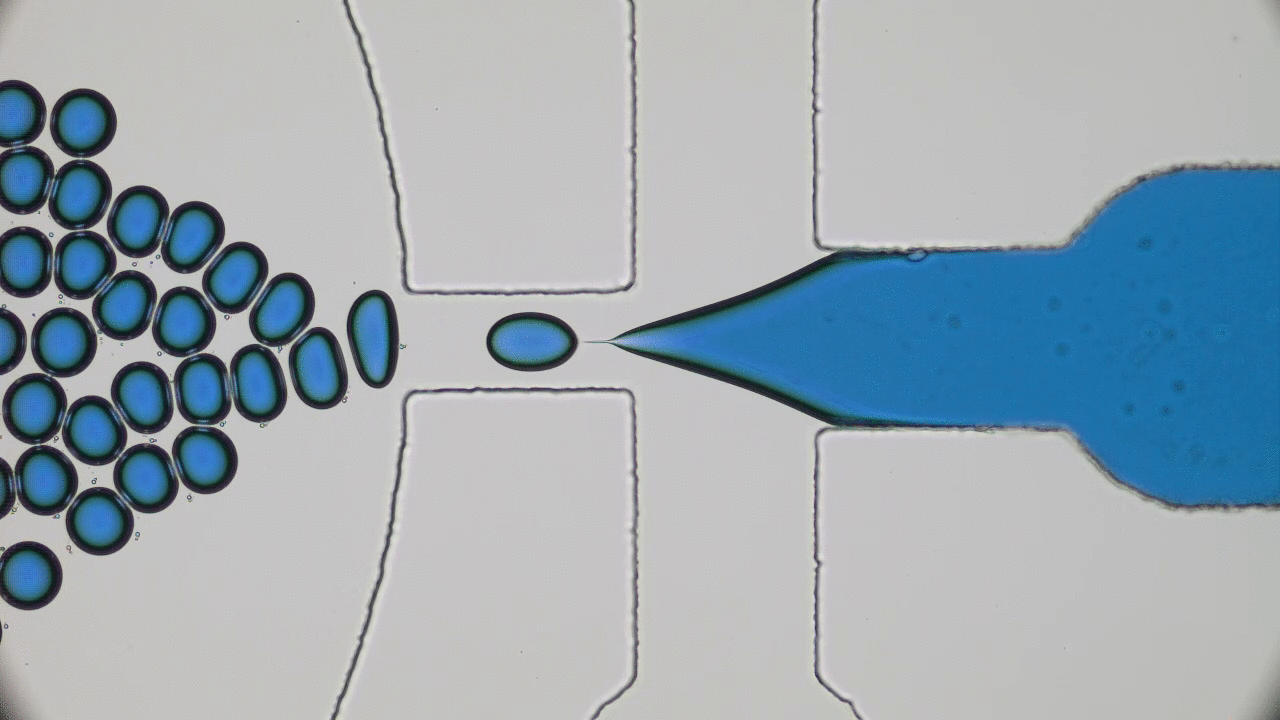
Brick-by-brick to Build Tiny Capsules
In past two decades, several approaches have been developed and optimized to encapsulate a wide variety of materials, from food to cosmetics and the more demanding realm of therapeutic reagents. Inspired by biological cells, the first attempts were to use either natural or synthetic lipid molecules to form encapsulation vessels, i.e., liposomes. Then, with the increasing awareness of controlled release of cargo, especially for therapeutic purposes, advanced materials such as polymers were developed to form carrying vessels. Despite the enormous progress in encapsulation technologies, however, these methods can be limited in their applicability regarding encapsulation efficacy, permeability, mechanical strength, and for biological applications, compatibility.