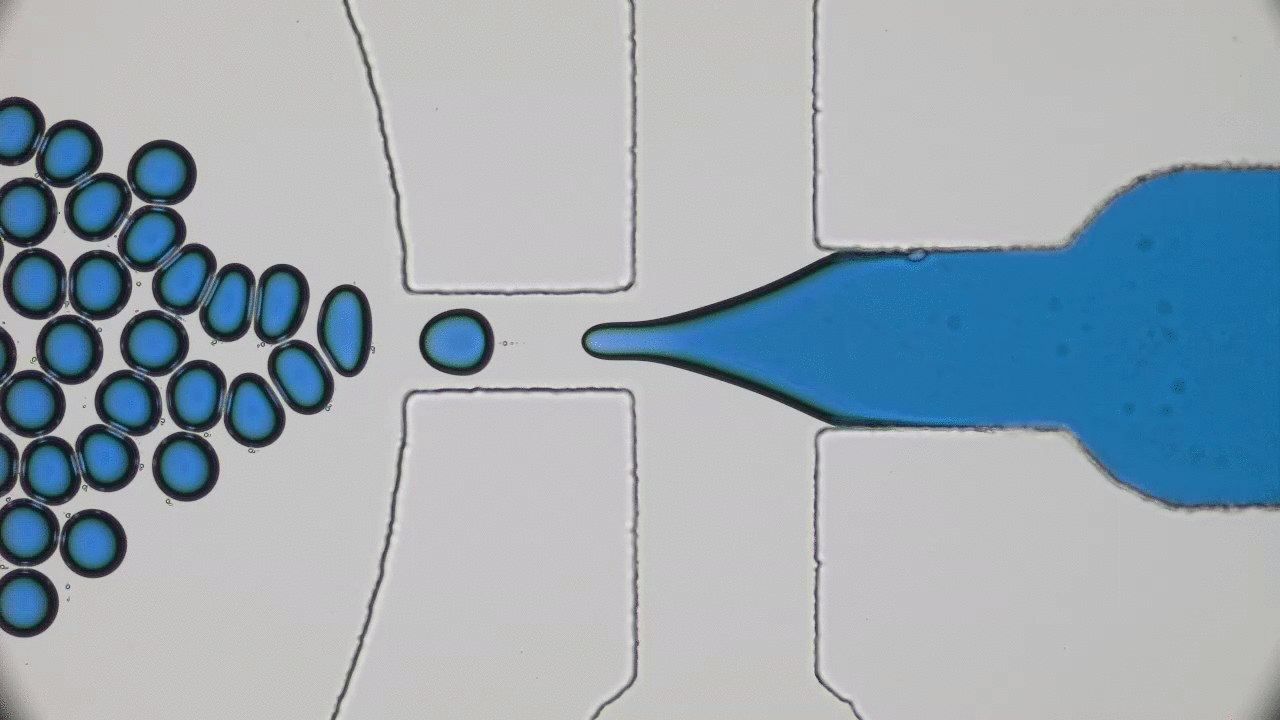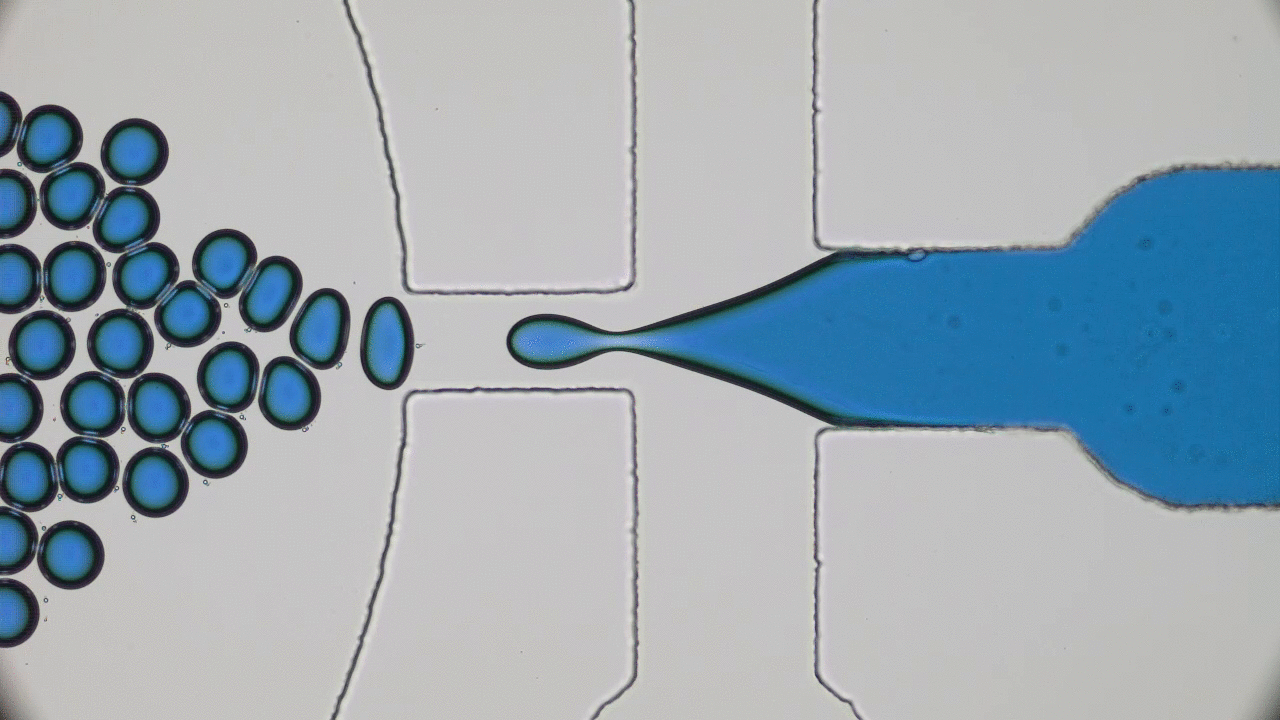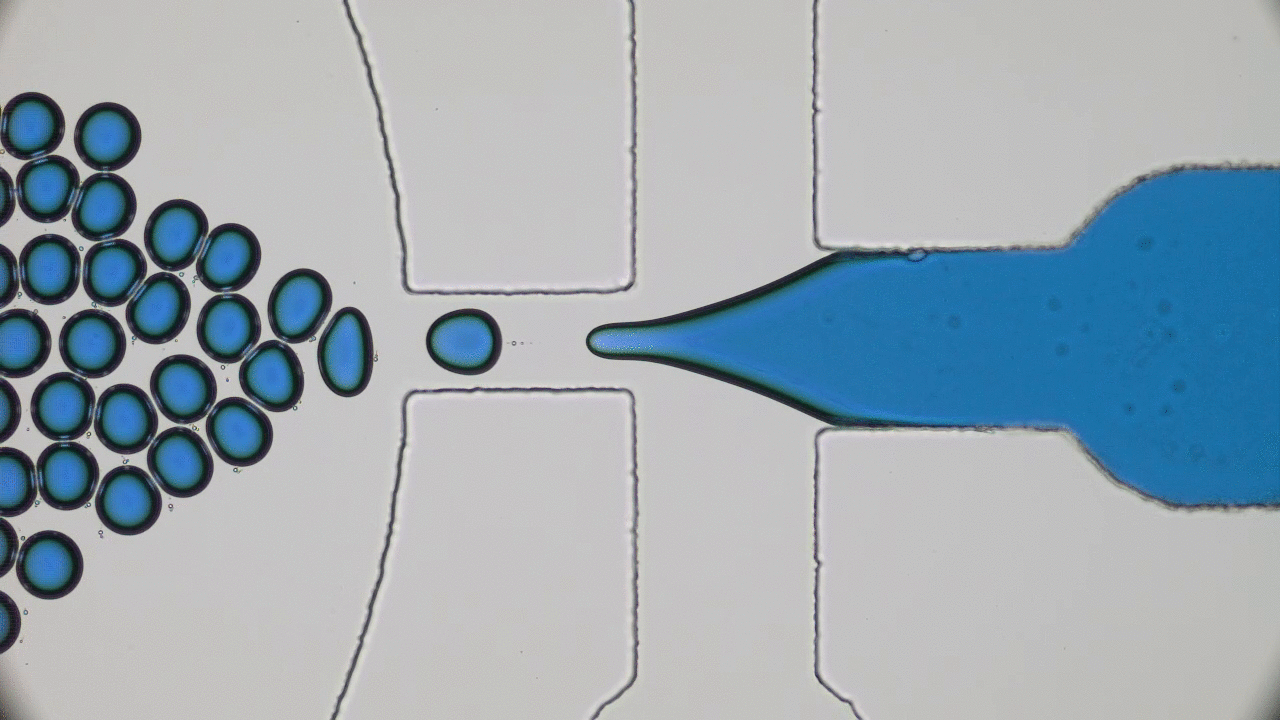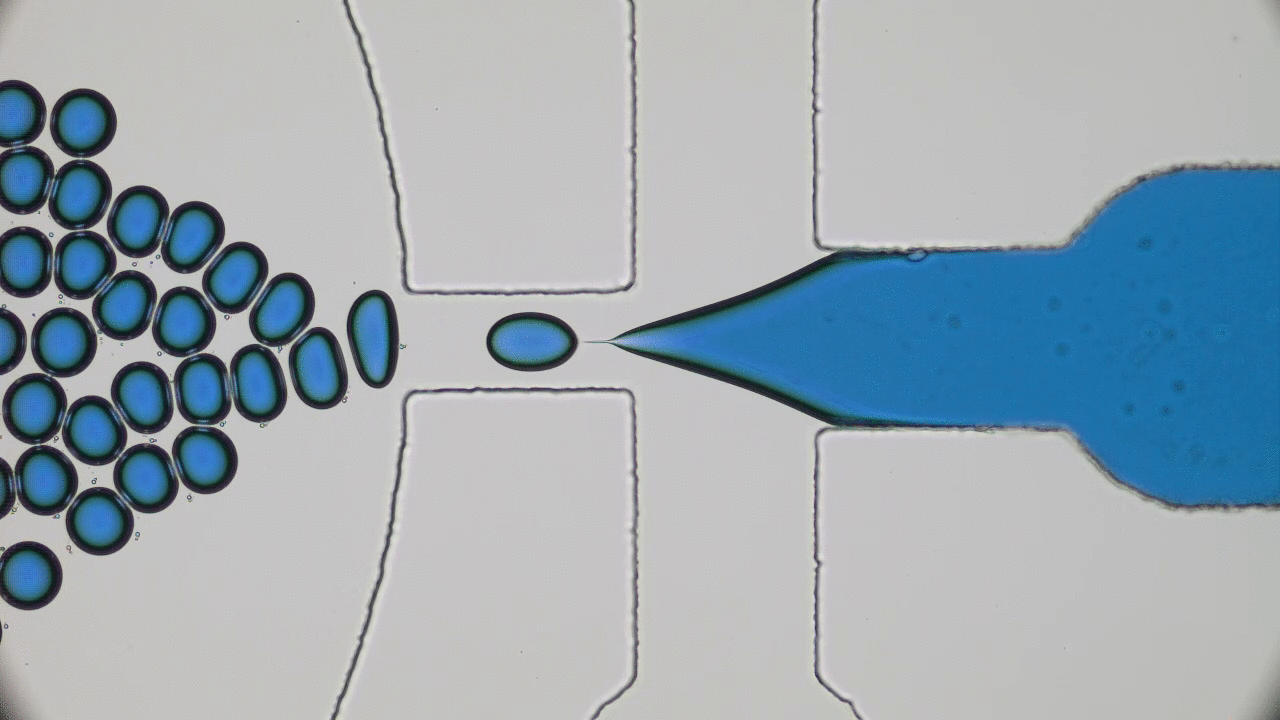
Imagine you and your friends are trapped by a super-villain in a cage. There is a giant gear with a diameter half the length of a football field in the center. The only way to open the cage door, get out, and stop the villain’s evil plans will be to rotate this gear by one full revolution. This is a daunting task for one person — but if you have enough friends, you can grab the gear’s teeth and push it together to escape. An analogous task is faced by flocks of tiny bacteria in today’s two featured papers. In “Bacterial ratchet motors”, Di Leonardo and colleagues discuss the mechanics of bacteria pushing a single gear, and in “Swimming bacteria power microscopic gears”, Sokolov and colleagues discuss how bacteria can interact to power more than one gear.
Anti-biofilm Material to Fight Bacterial Formation on Surfaces
Biofilms cause health problems for millions of people worldwide every year, primarily because of infections during surgery or consumption of contaminated packaged foods. To prevent these problems, some scientists are developing surface coatings that will prevent biofilm formation in the first place. In this week’s paper, we will learn about a new technique for creating a microscopic “shield” against the formation and growth of biofilms.
Coil and Recoil: New screw-like bacteria swimming
No one likes being stuck. Whether you are in a car stranded in mud or stuck in a dead-end job, continuing normal behaviour is unlikely to help. Whereas we can see approaching hazards and dead-ends and try to avoid them, bacteria must blindly swim through passageways and channels that are of a similar size to themselves, often resulting in the cell becoming trapped. So, how does a bacterium change its behaviour to free itself?
How swimming bacteria spin fluid
In today’s study, Dunkel and his colleagues investigate how bacteria can make flow patterns that look turbulent – chaotic and full of vortices – even though bacteria are tiny and slow. The bacteria push the fluid around as they swim and create vortices, spinning regions in the fluid. The 5 ?m long bacteria create vortices with diameters of 80 ?m by swimming at the speed of 30 ?m/s!