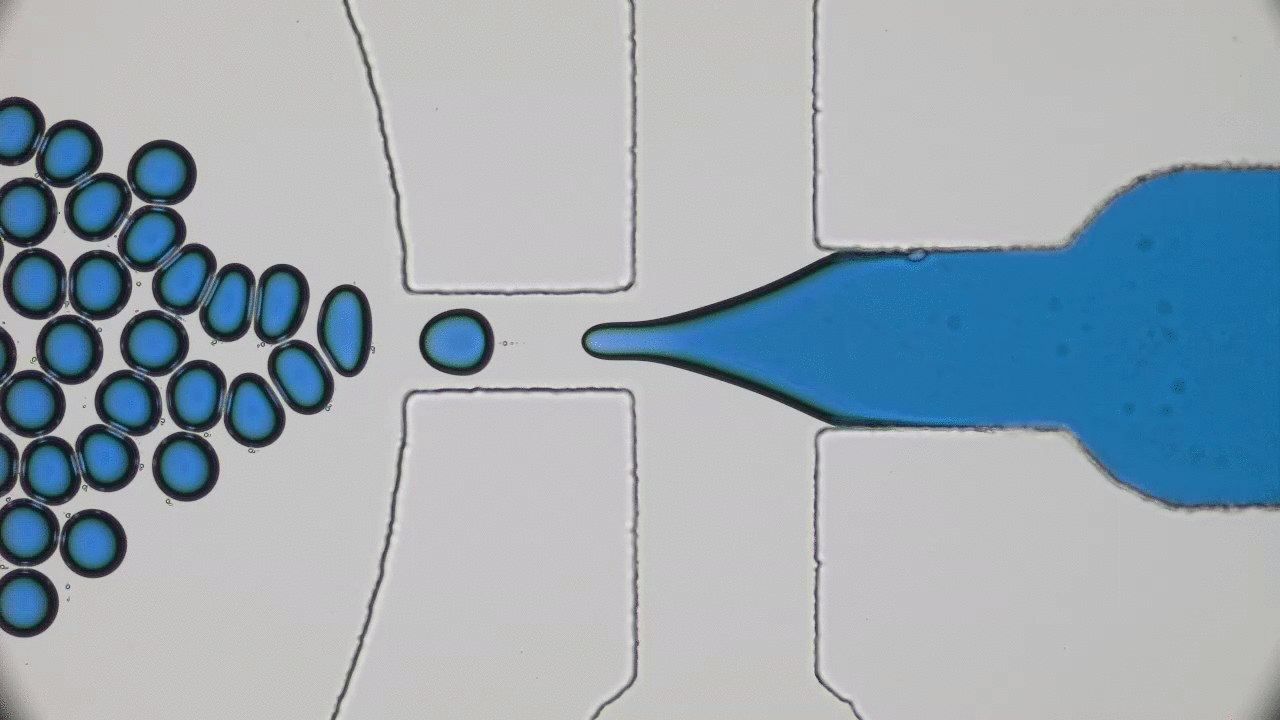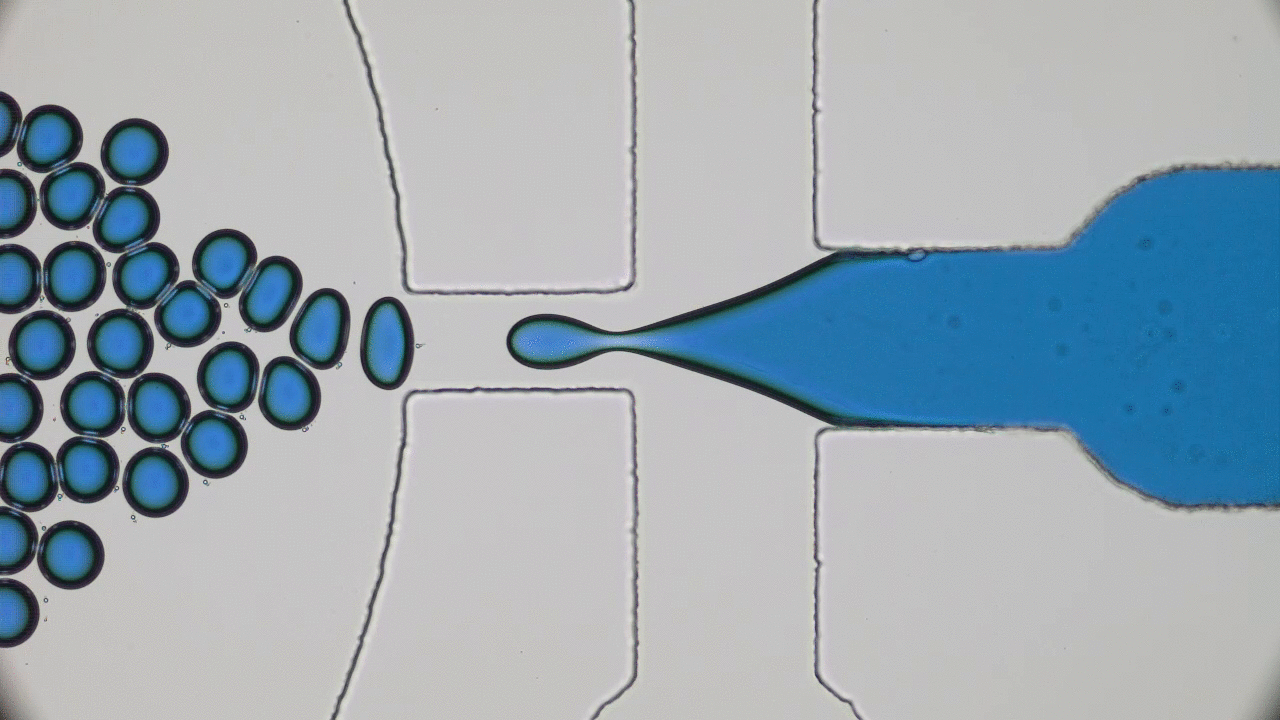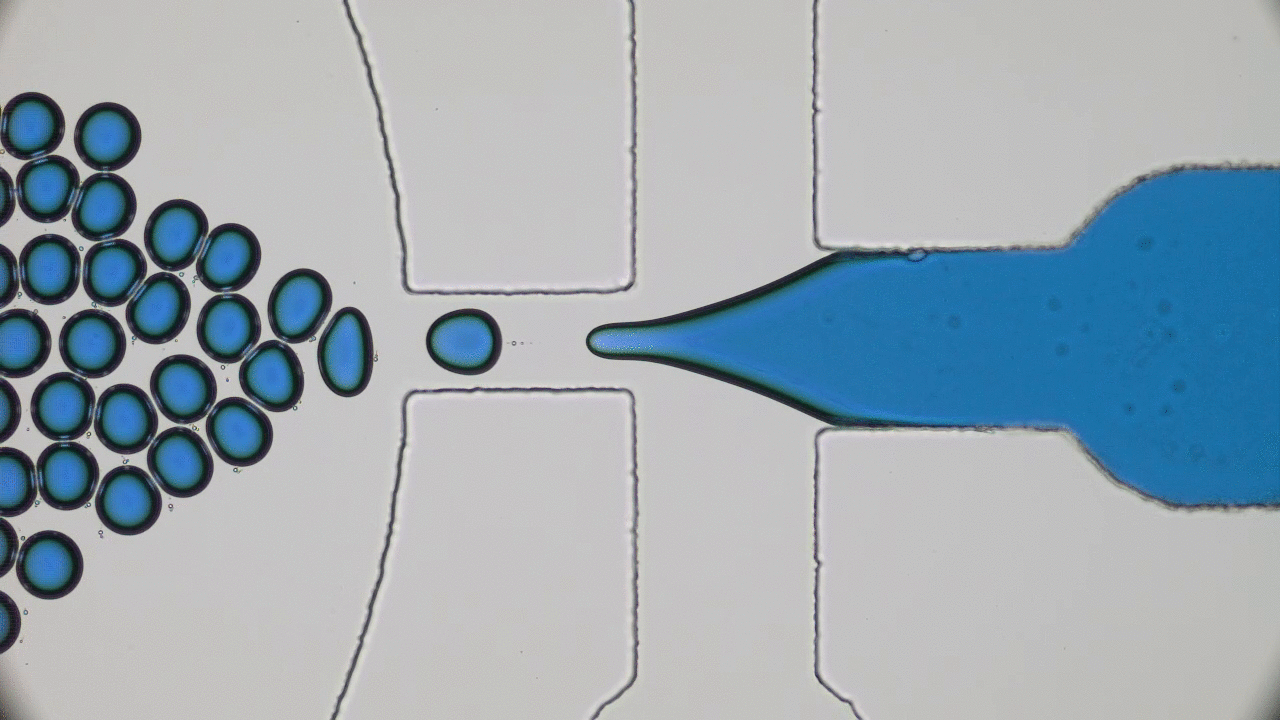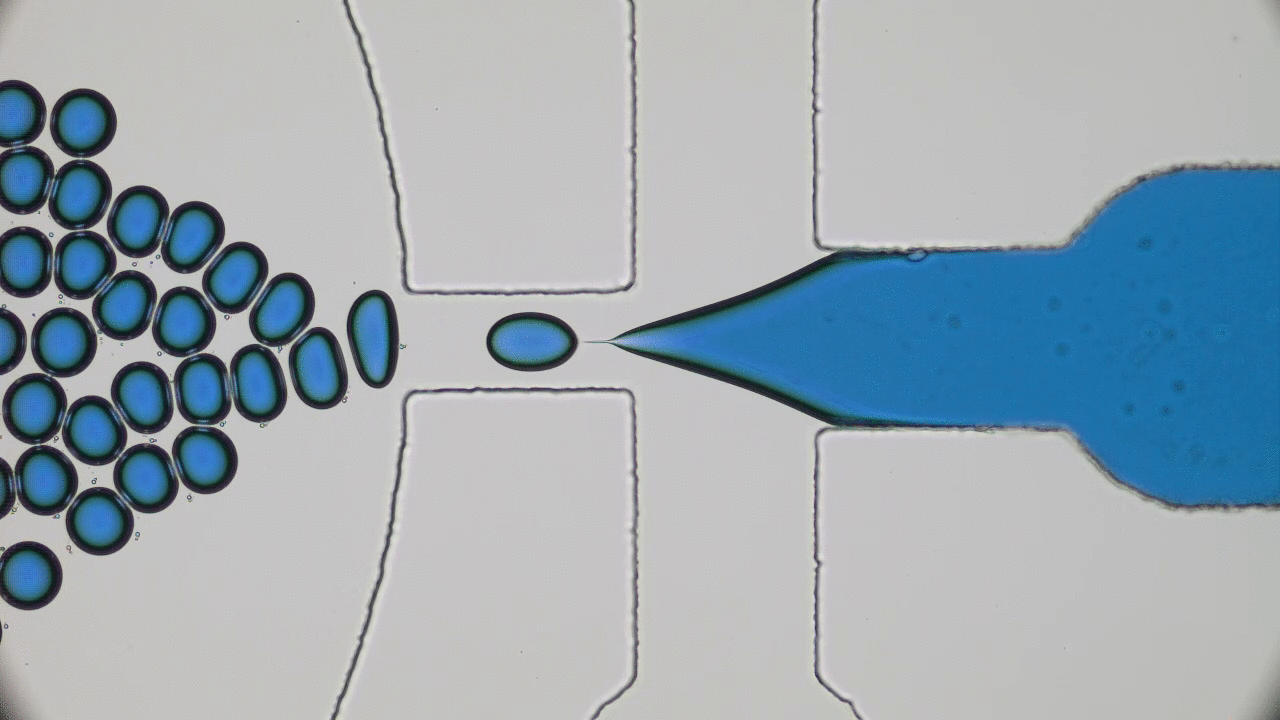
Quintessential soft matter problems, such as the behavior of droplets in ink-jet printing, involve complex interactions between forces and materials. In today’s article, Prof. Wilson Poon points out that coronaviruses are also quintessential soft matter objects, and highlights a range of areas where soft matter science may help better understand, and combat viral pandemics.
Who needs polymer physics when you can get worms drunk instead?
If you speak to a soft matter physicist these days, within a few minutes the term “active matter” is bound to come up. A material is considered “active” when it burns energy to produce work, just like all sorts of molecular motors, proteins, and enzymes do inside your body. In this study, the scientists are focusing specifically on active polymers. These are long molecules which can burn energy to do physical work. Much of biological active matter is in the form of polymers (DNA or actin-myosin systems for example), and understanding them better would give direct insight into biophysics of all kinds. But polymers are microscopic objects with complex interactions, making them difficult to manipulate directly. To make matters worse, physicists have yet to fundamentally understand the behaviors of active materials, since they do not fit into our existing theories of so-called “passive” systems. In this study, Deblais and colleagues decided to entirely circumvent this problem by working with a much larger and easier-to-study system that behaves similarly to a polymer solution: a mixture of squirming worms in water.
The trajectories of pointy intruders in sand
An alien spaceship commander was preparing to drop a cone-shaped spy shuttle into the sand of a Florida beach near Cape Canaveral. The shuttle needed to burrow deep enough that any passing humans wouldn’t see it while the aliens used it to spy on Earth’s space program. “From how high should I drop the shuttle so that it is hidden?” the commander asked their science advisor. The science advisor pulled out their alien high school mechanics book, hoping to calculate this based on the laws of motion and Earth’s gravitational force.
Supersoft matter bounces back: Softer, Better, Stronger
If you’ve ever worn soft contact lenses, you may know that they dry out and harden if they are not stored in a solution. This pervasive issue of hydrogel materials occurs when the solvent leaks or evaporates, affecting their mechanical properties. In this week’s post, polymer scientists develop super-soft dry elastomers (very elastic or rubbery polymers) that surpass the softness and elasticity of hydrogels, all without getting their hands wet.
For the People, By the People: Early career researchers organize virtual polymer physics symposium
In these unprecedented and fluid times, conferences and symposia have gone virtual as STEM collectively settles into a new normal. Many large meetings, like the formerly “in-person only” American Physical Society (APS) and American Chemical Society (ACS) national meetings, have been cancelled or transitioned to virtual-only participation this year. The 2021 Spring APS meeting will go virtual as well. I love big in-person meetings and have shied away from virtual alternatives thinking they would not provide the same feeling of community with my fellow scientists. However, the isolation of quarantine and the desire to get comfortable with the “new normal” motivated me to step out of my comfort zone and into the world of virtual science meetings this summer. So, when the opportunity to attend the 2020 Virtual Polymer Physics Symposium (VPPS) arose in July, I jumped at the chance to participate.
Lifehack: How to pack two meters of chromatin into your cell’s nucleus, knot-free!
The entirety of our genetic information is encoded in our DNA. In our cells, it wraps together with proteins to form a flexible fiber about 2 metres long known as chromatin. Despite its length, each cell in our body keeps a copy of our chromatin in its nucleus, which is only about 10 microns across. For scale, if the nucleus was the size of a basketball, its chromatin would be about 90 miles long. How can it all fit in there? To make matters worse, the cell needs chromatin to be easily accessible for reading and copying, so it can’t be all tangled up. It’s not surprising then that scientists have been puzzled as to how this packing problem can be reliably solved in every cell. The solution is to pack the chromatin in a specific way, and research suggests that this may be in the form of a “fractal globule”.
APS March Meeting self-organizes online
I was ready. I was so ready. I had all my chargers and AV adapters. My presentation was backed up on a USB drive. I had every talk I wanted to go to on my calendar. I had sent emails to professors I wanted to meet and network with. I reached out to friends I only see in March in a different city every year. It was 10 pm on Saturday, February 29. My flight to Denver was leaving at 6 am in the morning. Then the email arrived —
How to turn off cancer cell growth using mechanosensing
Our bodies are made up of cells that can sense and respond to their dynamic environment. As an example, pancreatic beta cells chemically sense increased blood sugar concentrations and respond by producing insulin. Scientists have found that cells can also mechanically sense their environment; “mechanosensing” determines whether a cell should grow or die. Cancer is characterized by uncontrolled cellular growth, where cells often contain mutations that inhibit the natural mechanisms of cell death. Because mechanosensing is one such mechanism, scientists have hypothesized that cancer cells keep growing because they lack the ability to probe their environments. In this week’s paper, published in Nature Materials, an international research team led by Bo Yang and Michael Sheetz from the National University of Singapore investigated that hypothesis by combining tools from soft matter physics and biology.
Researchers play with elastic bands to understand DNA and protein structures.
Recently, Nicholas Charles and researchers from Harvard published a study that used simulations of elastic fibers to probe their response to stretching and rotation applied simultaneously. The results shed light on how DNA, proteins, and other fibrous materials respond to forces and get their intricate shapes.
Huddling penguins make waves in the Antarctic winter
Standing in the center of a crowded bus on your way to class, you might think: “why don’t these people just move? It’s hot and I can’t breathe!” Male penguins huddling to keep their eggs warm in the Antarctic winter have the opposite problem – no penguin wants to be at the cold edge of the huddle. A penguin in the huddle wants to stay in the warm center, since the outside temperature can reach -45 oC. However, penguins on the edge of the huddle are trying to push through the crowd to reach the center. Through the independent motion of each penguin, the huddle stays tight enough for the center to remain warm but loose enough to keep moving.