I was ready. I was so ready. I had all my chargers and AV adapters. My presentation was backed up on a USB drive. I had every talk I wanted to go to on my calendar. I had sent emails to professors I wanted to meet and network with. I reached out to friends I only see in March in a different city every year. It was 10 pm on Saturday, February 29. My flight to Denver was leaving at 6 am in the morning. Then the email arrived —
Spell Checking Boiolgy
Cells have to do an awful lot of tasks correctly and on time in a noisy and unpredictable environment. How do they do it? In this post, we look at one particular process, making proteins, and learn how cells operate away from thermodynamic equilibrium in order to make the right protein at the right time over and over again.
Scaling up biology
The difference between a bacterium and a whale are huge, and not just their size. However, there are hidden scaling laws underlying all living things. These scaling laws are found to be due to the fractal-like nutrient distribution systems. Here, we review how to derive the scaling law for metabolic rate with organism mass, illustrating its generality and ubiquity.
How the leopard got its spots
Patterns are ubiquitous in nature, but how do they form? By considering how proteins can interact with each other, Alan Turing gives an explanation of how patterns can be formed without any genetic control, simply by following known laws of physics.
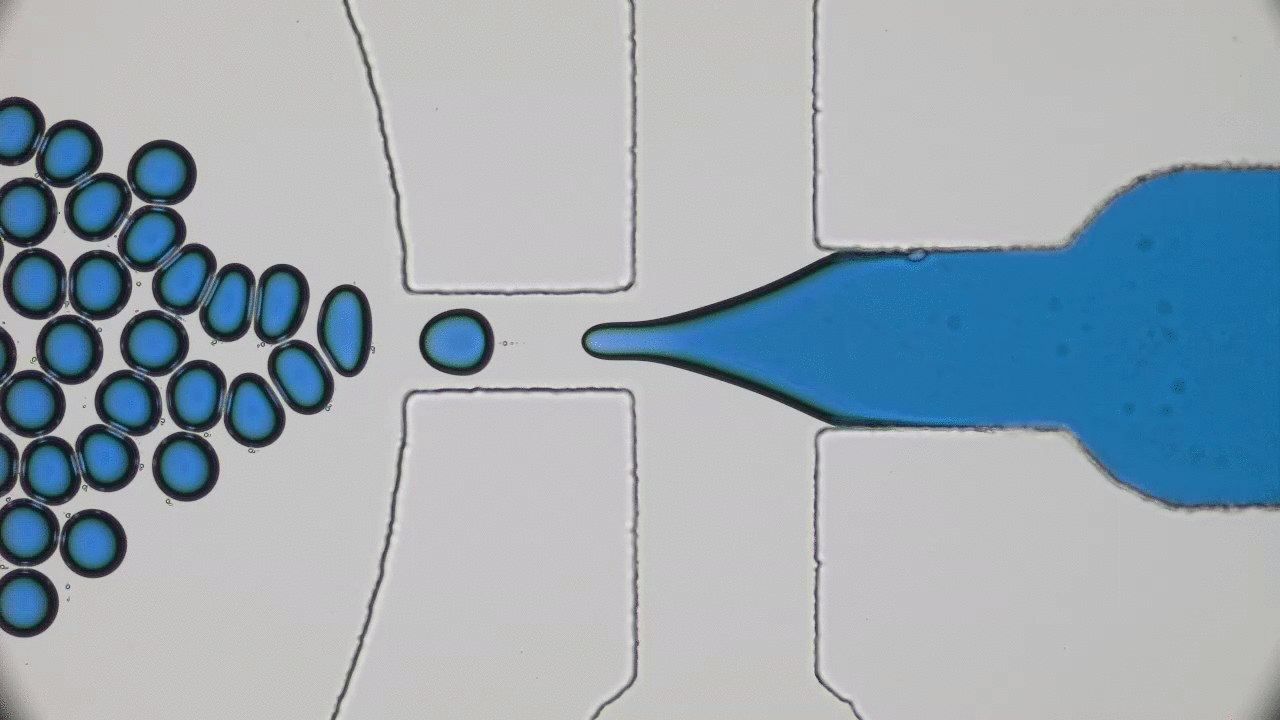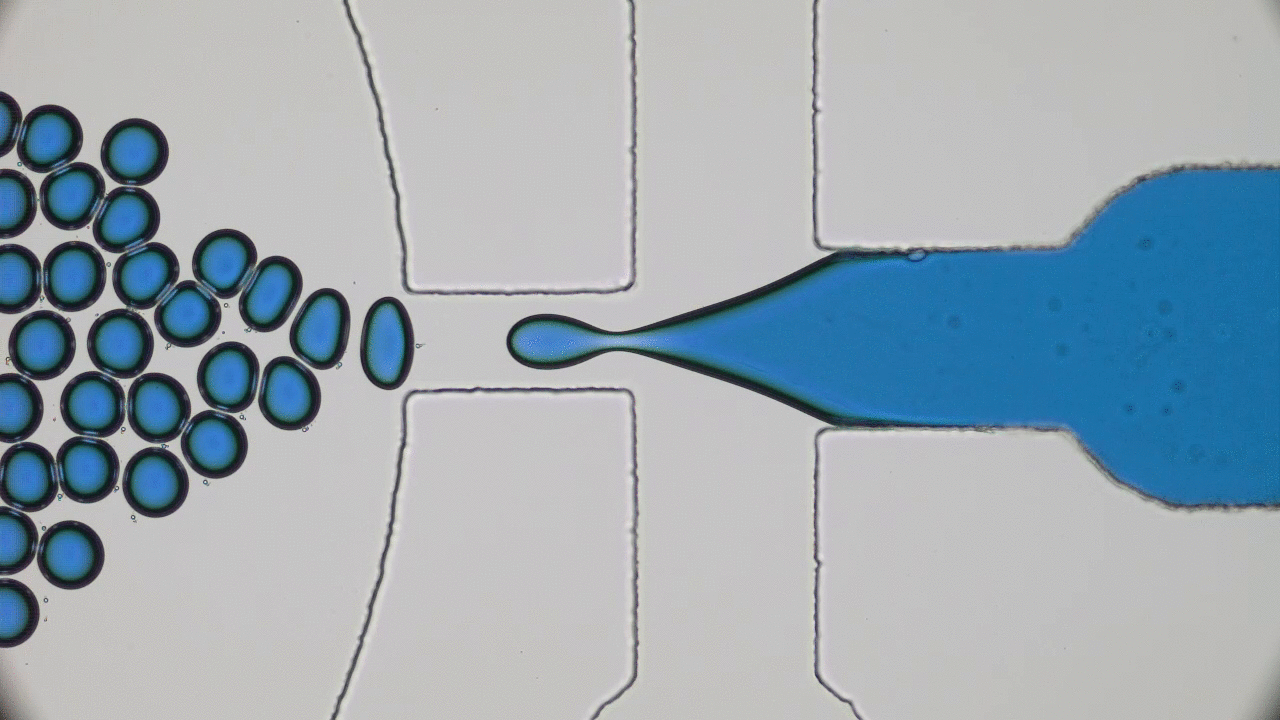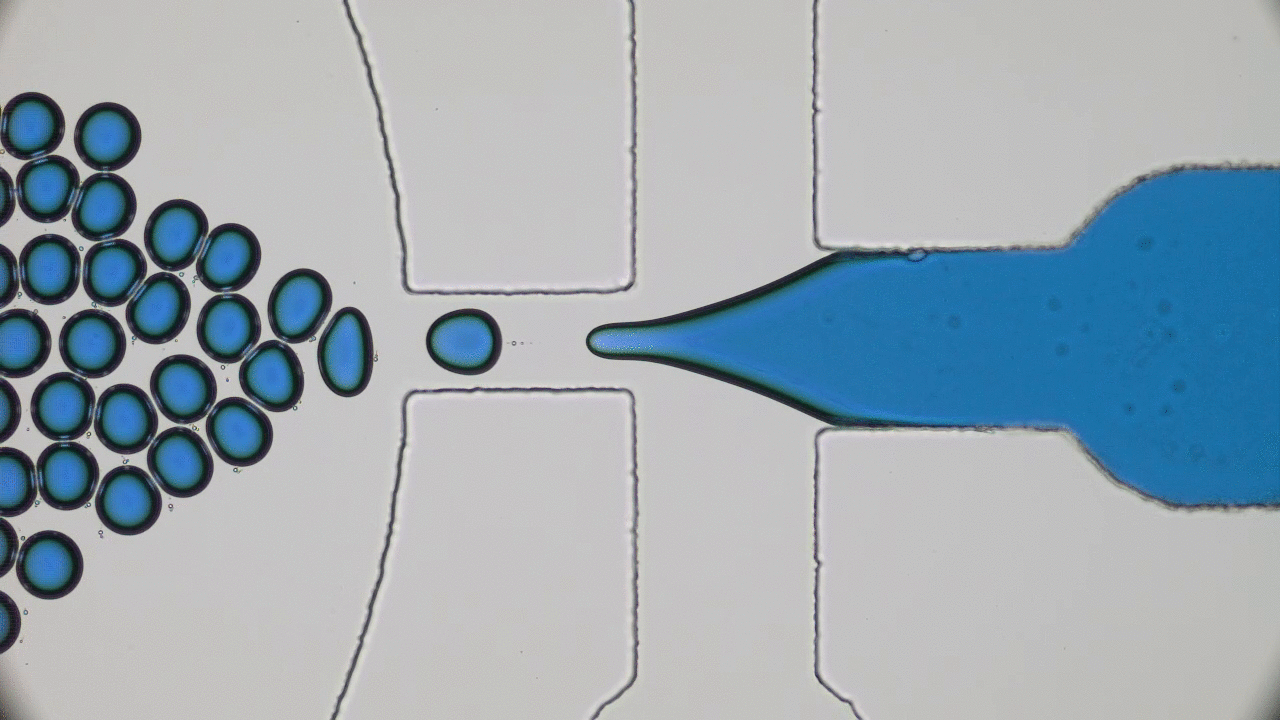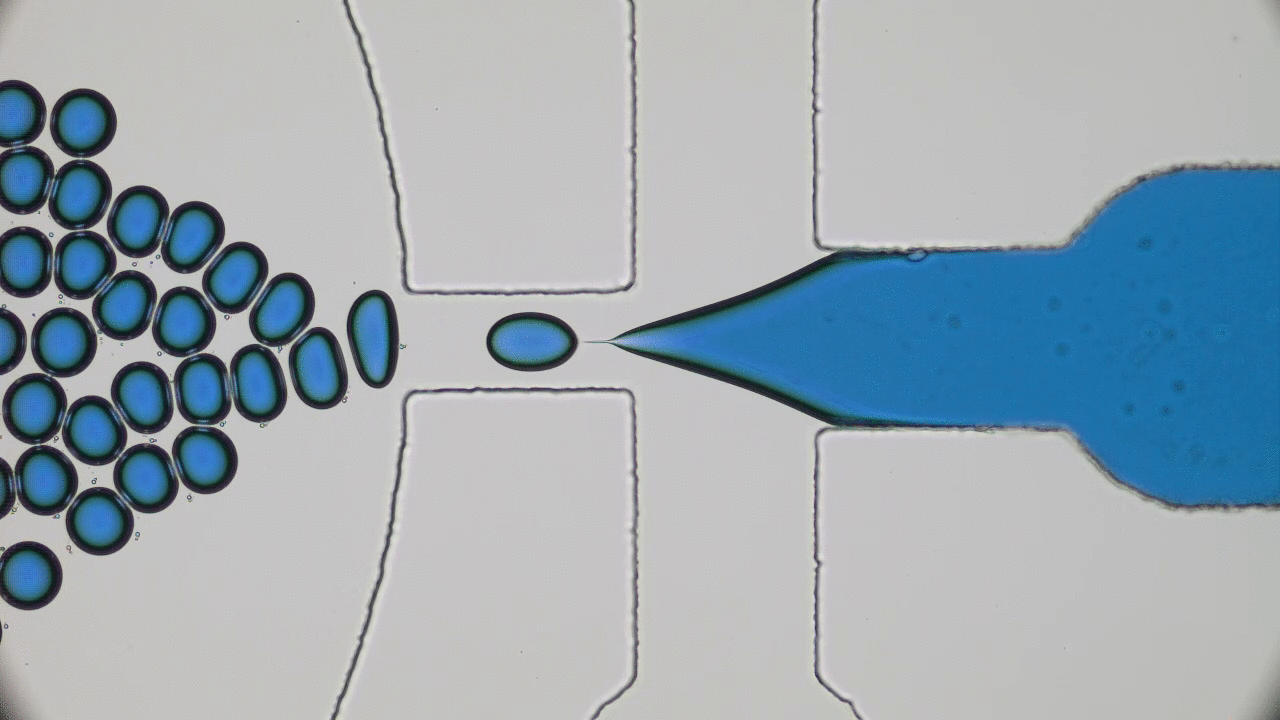
Dividing Liquid Droplets as Protocells
In the beginning there was… what, exactly? Uncovering the origins of life is a notoriously difficult problem. When a researcher looks at a cell today, they sees the highly-polished end product of millennia of evolution-driven engineering. In today’s paper, David Zwicker, Rabea Seyboldt, and their colleagues construct a relatively simple theoretical model for how liquid droplets can behave in remarkably life-life ways.