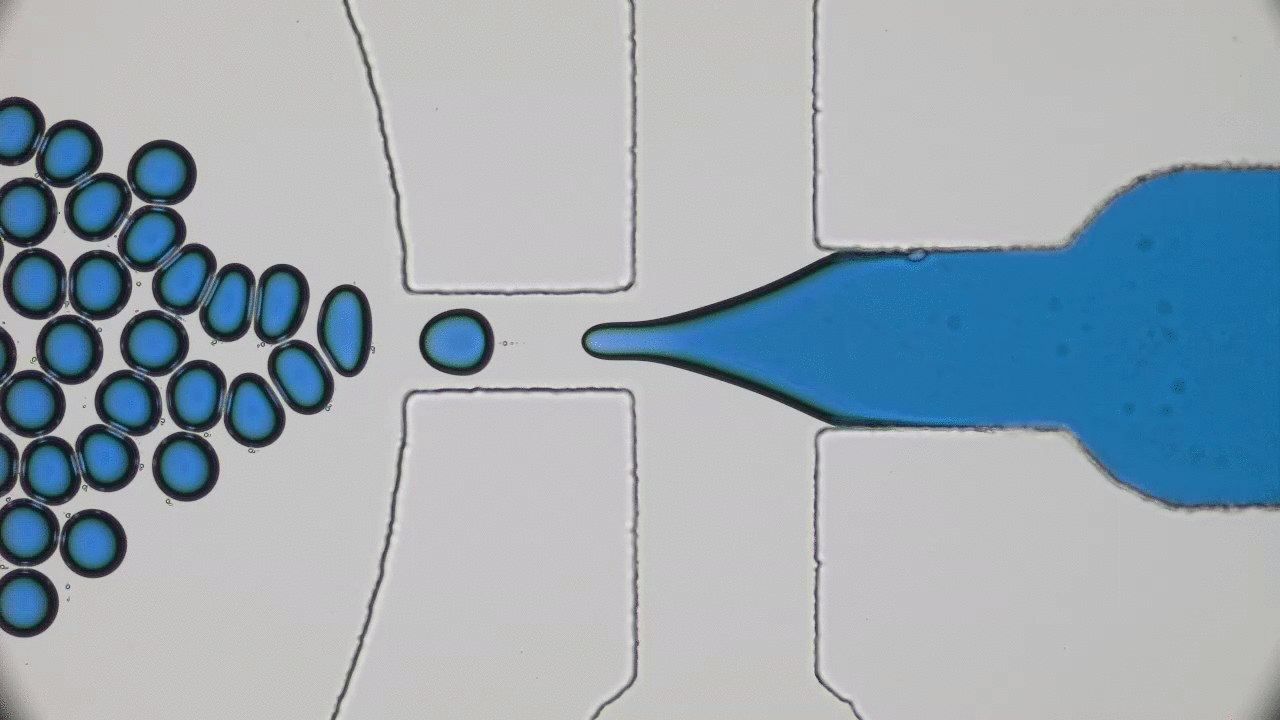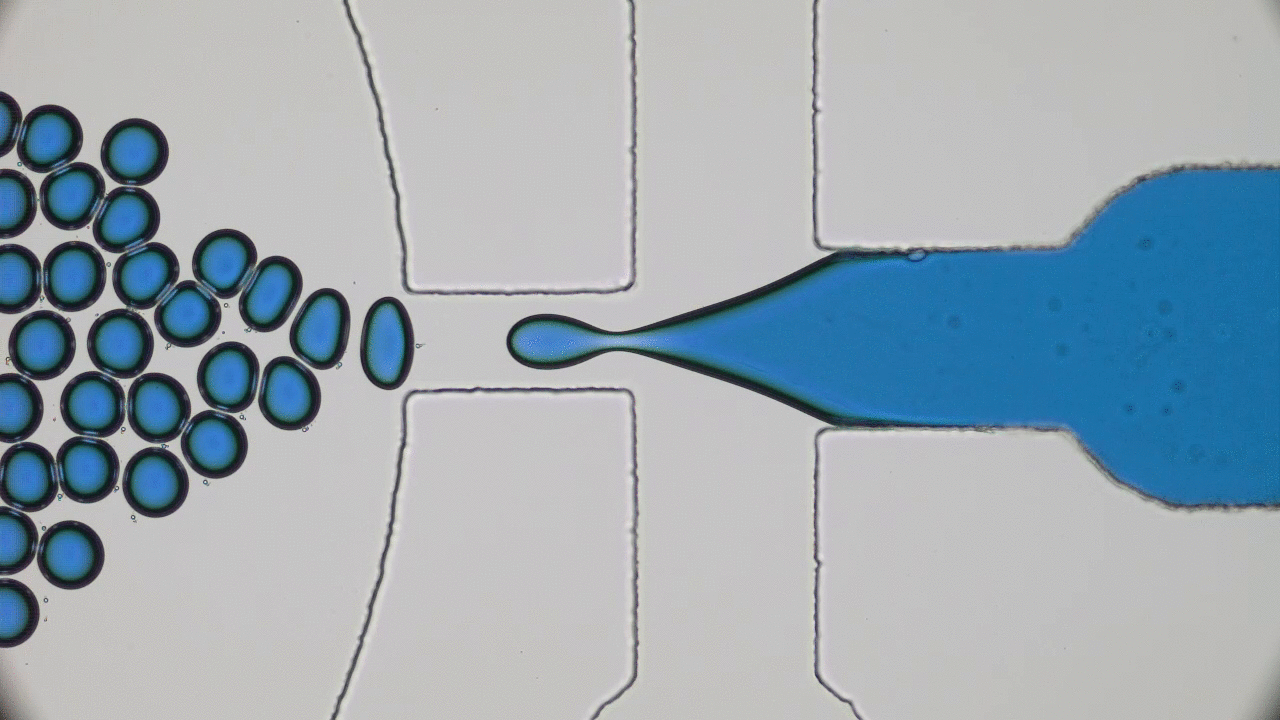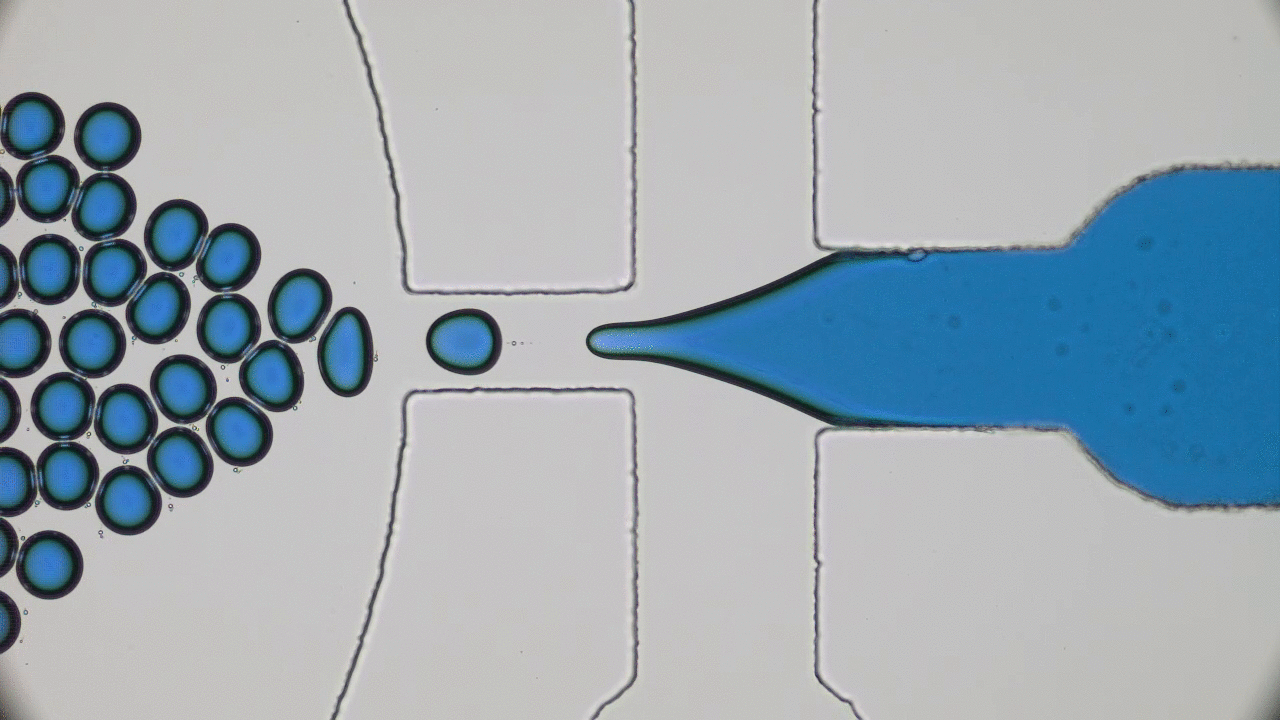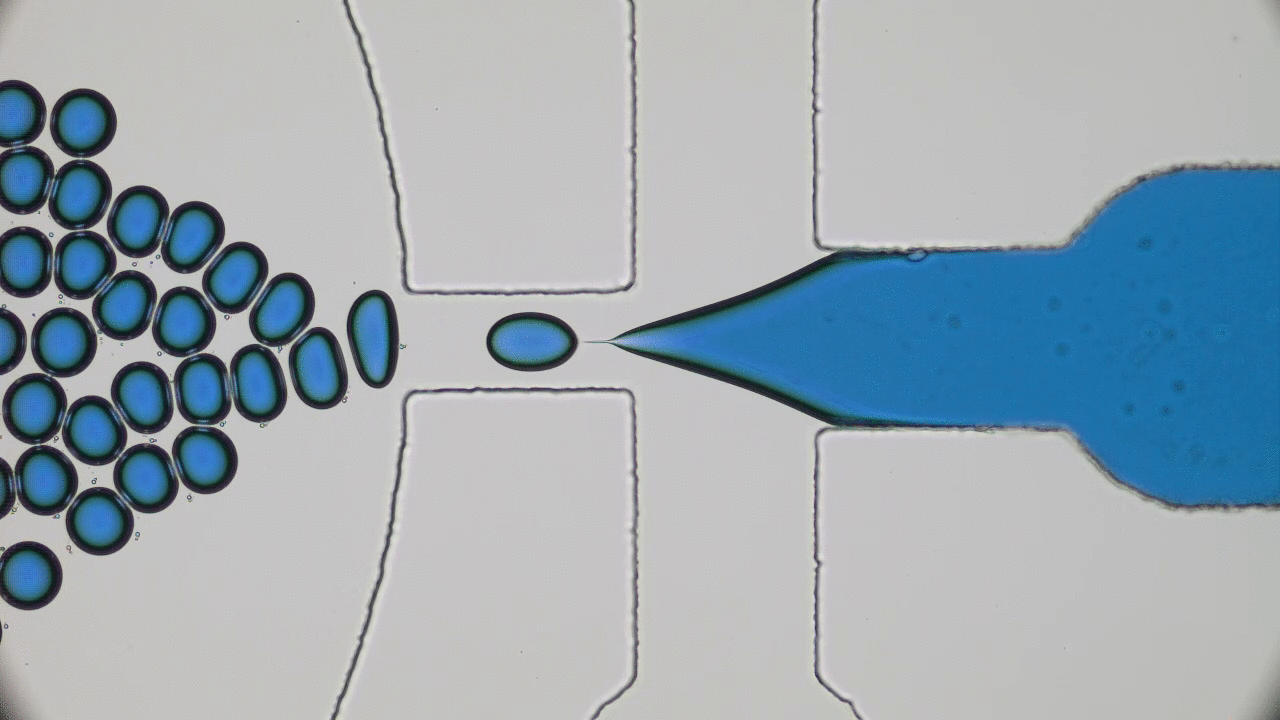
In the 1966 movie Fantastic Voyage, a submarine and its crew shrink to the size of a microbe in order to travel into the body of an escaped Soviet scientist and remove a blood clot in his brain. The film gave viewers a glimpse into a possible future where doctors could treat patients by going directly to the source of the problem instead of being limited by the inaccessibility of most parts of the human body. This dream of a tiny submarine that can be piloted through the human body to deliver medical care remains, even 50 years later, in the realm of science fiction. However, Miskin and coworkers at Cornell University have brought us one step closer to making this a reality with their recent development of autonomous microscale machines.
Microcannons firing Nanobullets
Sometimes I read papers that enhance my understanding of how the universe works, and sometimes I read papers about fundamental research leading to promising new technologies. Occasionally though, I read a paper that is just inherently cool. The paper by Fernando Soto, Aida Martin, and friends in ACS Nano, titled “Acoustic Microcannons: Toward Advanced Microballistics” is such a paper.
Knotty DNA
Try taking out your earphones from your pocket and, in all probability, you’ll find knots and entanglements between the ends. As it turns out, this knotting effect is not limited to macroscopic objects, but occurs on the nanoscale as well. A DNA molecule that carries the genetic information of a living organism is actually a long string-like polymer, so you can imagine that it would also get tangled up just like the cords of your earphones. In today’s paper, Calin Plesa and his colleagues at TU Delft are able to observe and measure these knots in DNA strands and uncover behaviour which has not been observed before.