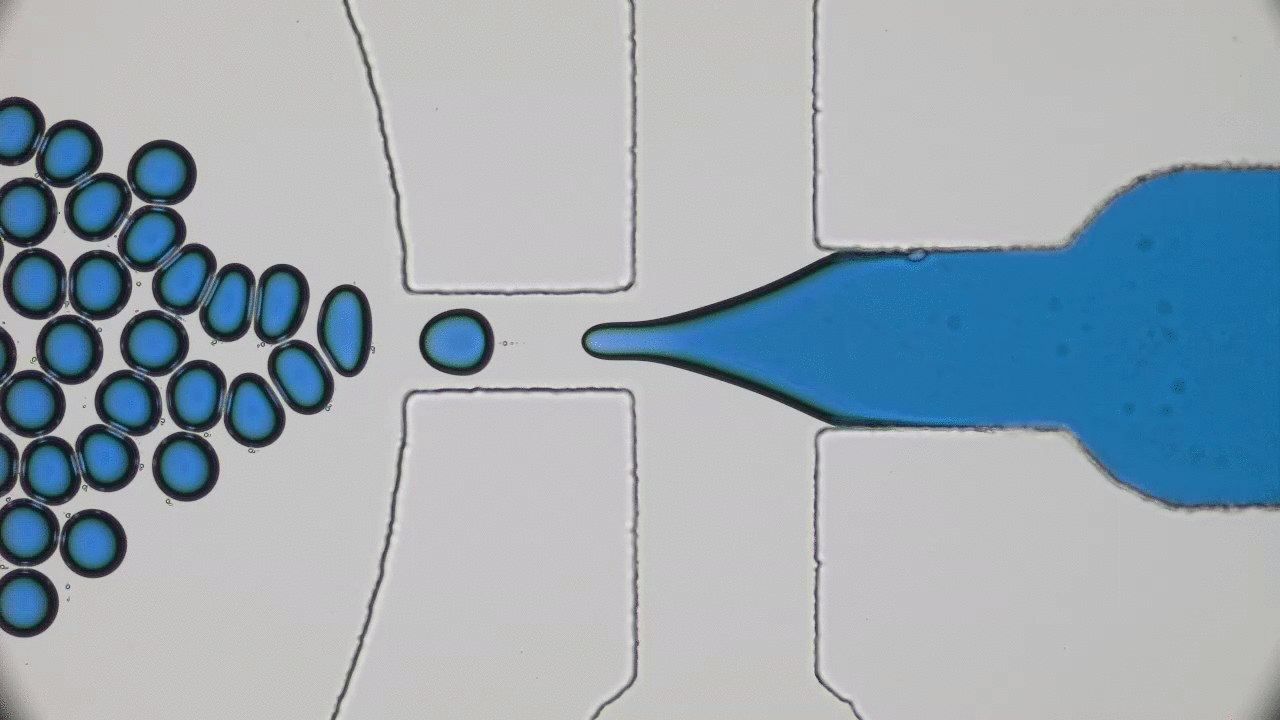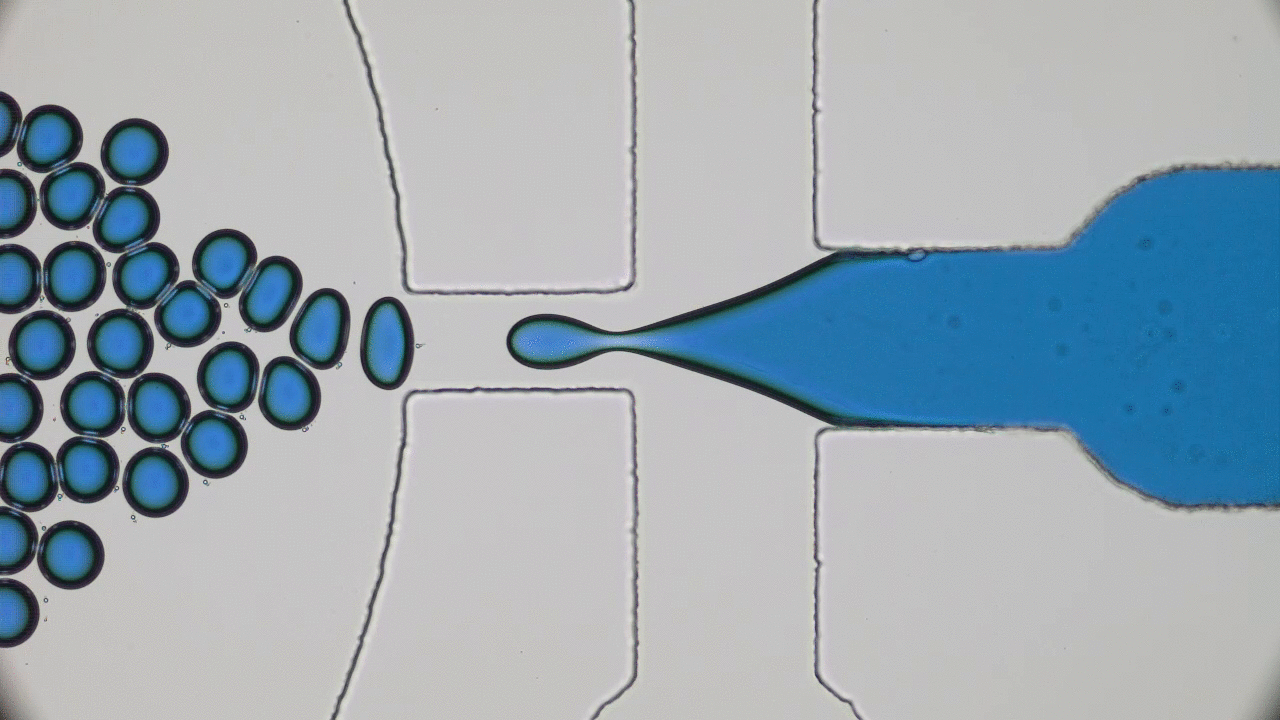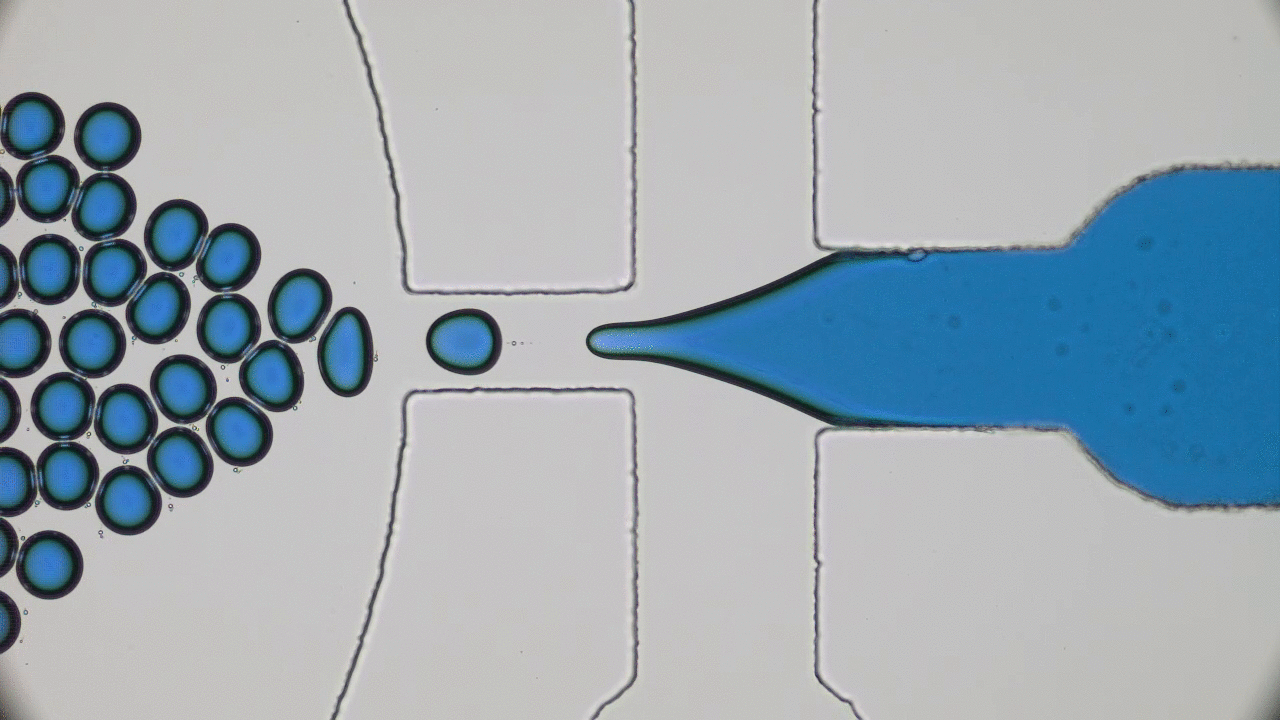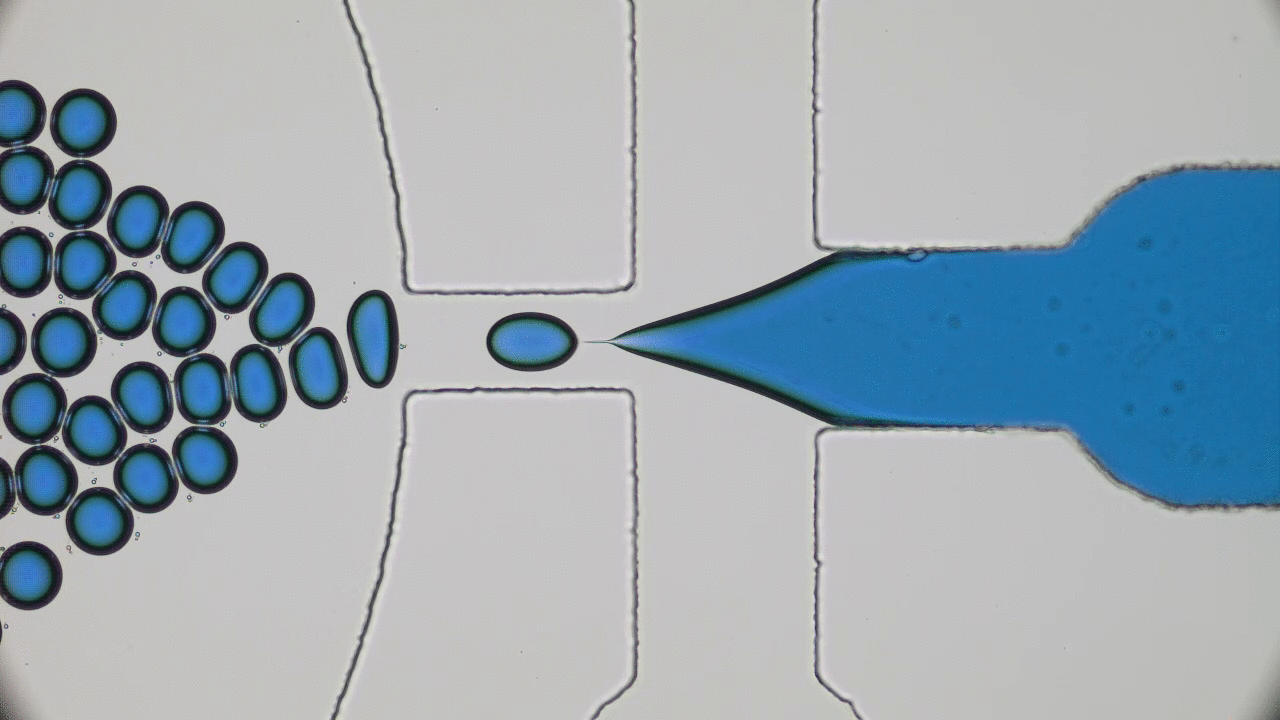
Understanding the origin of life is one of the most enduring and fundamental scientific challenges there is. Of all branches of science, physics is probably not the first place one would think to go to for enlightenment. Life seems too complicated and multi-layered to be captured by the simplistic frameworks of physics. Today’s paper tackles a small part of understanding the origin of life – the physics of self-replication.
Discovery of Liquid Crystals in Short DNA
Ever since its discovery, scientists have known that the DNA molecule is present in every life form. It carries the genetic information of all living organisms and many viruses. Today, however, we will strip DNA of its genetic importance and look at it from a different perspective. We will discuss why DNA attracts attention even outside of the biological context: What is the connection between DNA and liquid crystals? What are end-to-end stacking interactions and why are they important? If you want to get answers on these questions (and many more), keep reading.