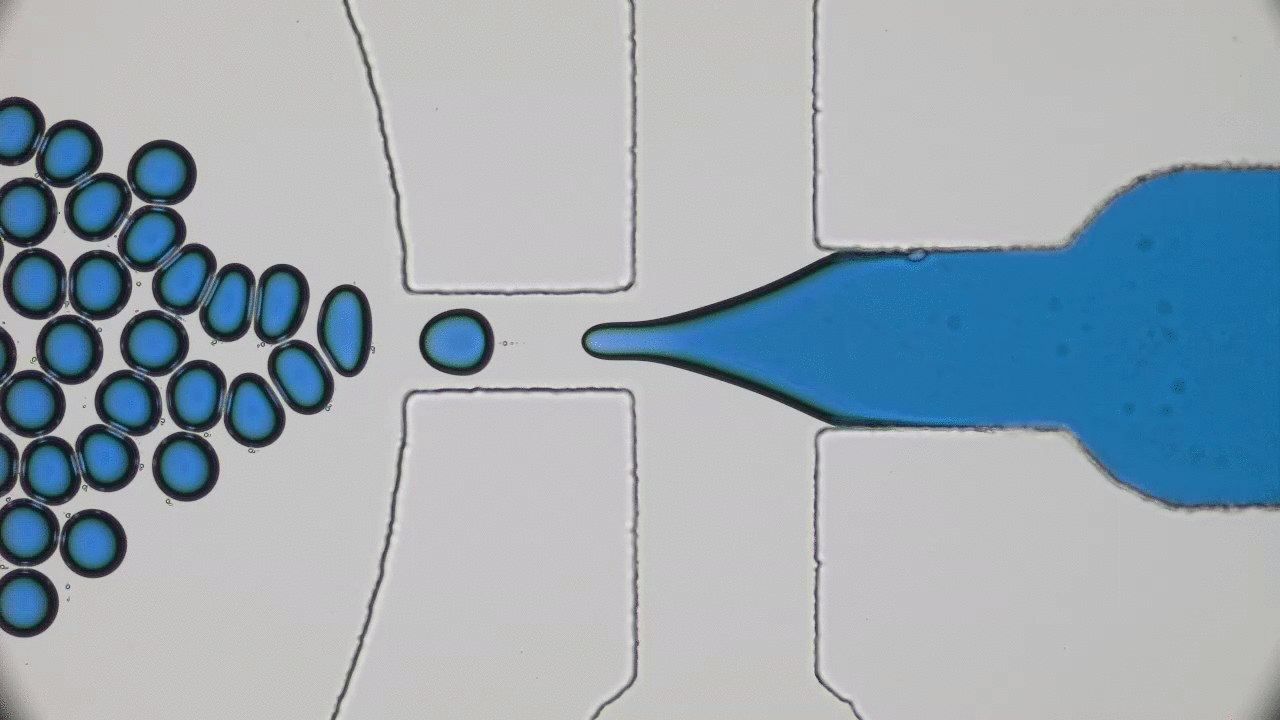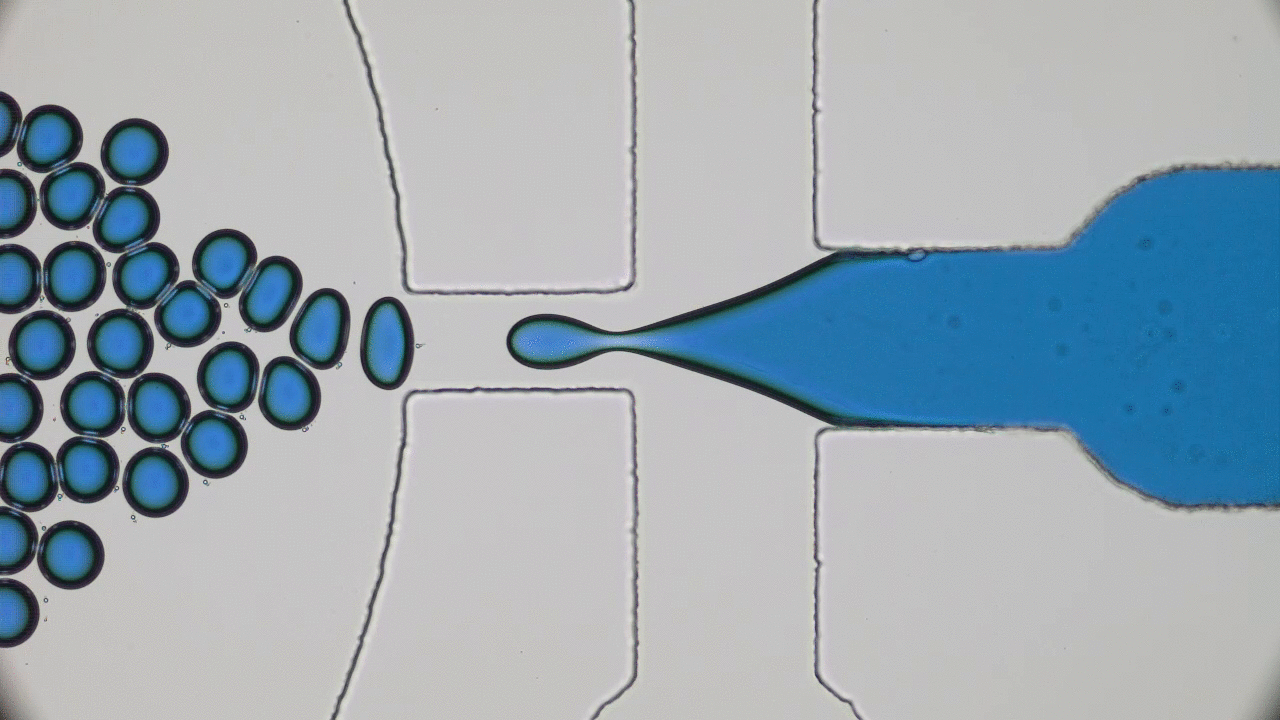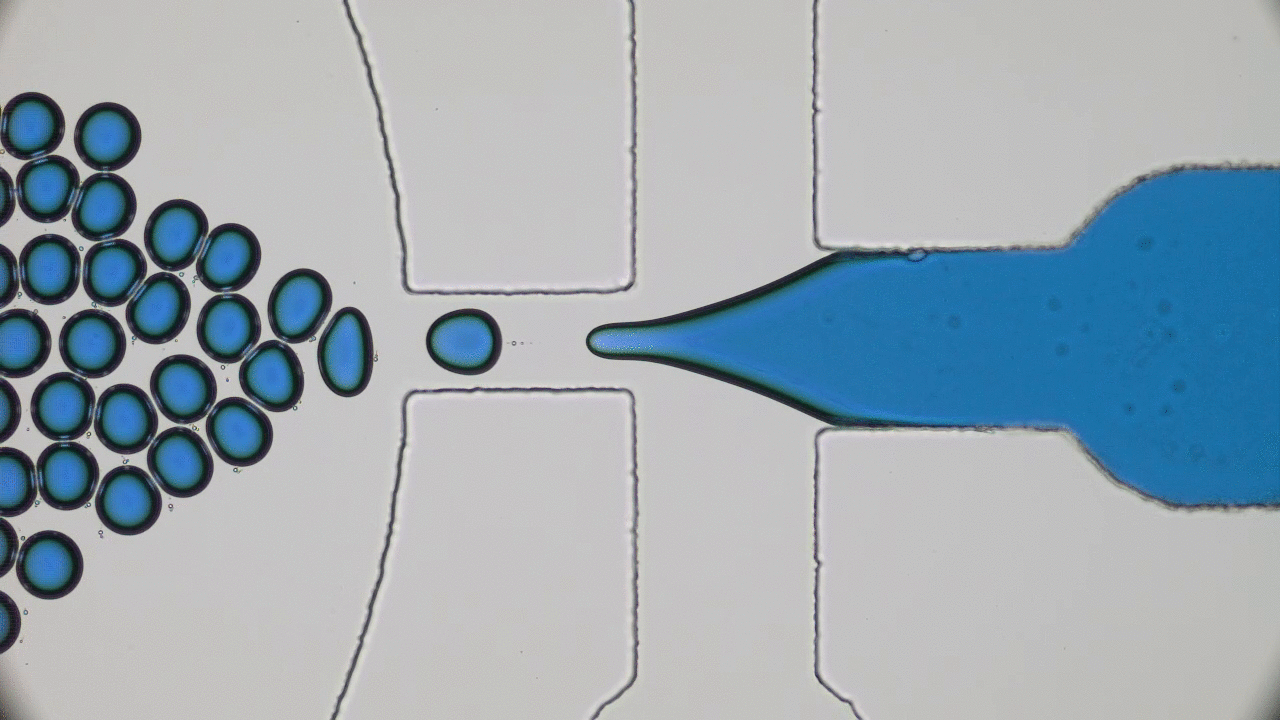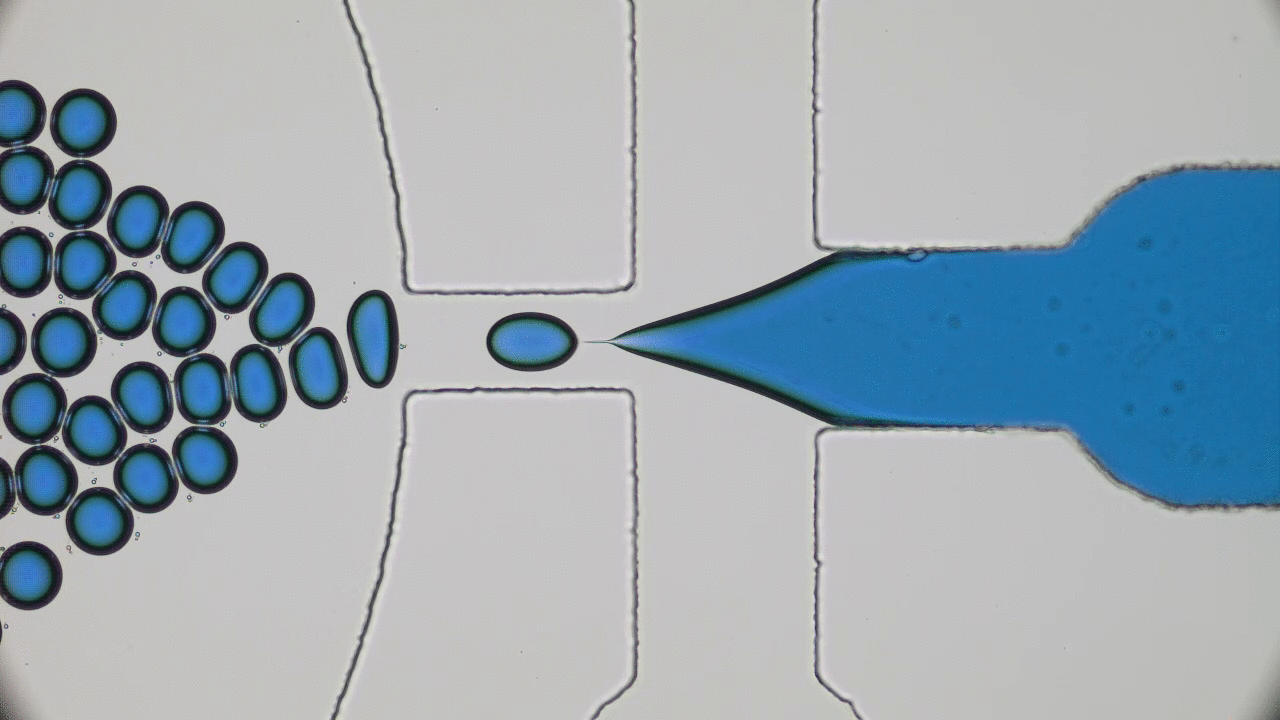
The entirety of our genetic information is encoded in our DNA. In our cells, it wraps together with proteins to form a flexible fiber about 2 metres long known as chromatin. Despite its length, each cell in our body keeps a copy of our chromatin in its nucleus, which is only about 10 microns across. For scale, if the nucleus was the size of a basketball, its chromatin would be about 90 miles long. How can it all fit in there? To make matters worse, the cell needs chromatin to be easily accessible for reading and copying, so it can’t be all tangled up. It’s not surprising then that scientists have been puzzled as to how this packing problem can be reliably solved in every cell. The solution is to pack the chromatin in a specific way, and research suggests that this may be in the form of a “fractal globule”.
Researchers play with elastic bands to understand DNA and protein structures.
Recently, Nicholas Charles and researchers from Harvard published a study that used simulations of elastic fibers to probe their response to stretching and rotation applied simultaneously. The results shed light on how DNA, proteins, and other fibrous materials respond to forces and get their intricate shapes.
Discovery of Liquid Crystals in Short DNA
Ever since its discovery, scientists have known that the DNA molecule is present in every life form. It carries the genetic information of all living organisms and many viruses. Today, however, we will strip DNA of its genetic importance and look at it from a different perspective. We will discuss why DNA attracts attention even outside of the biological context: What is the connection between DNA and liquid crystals? What are end-to-end stacking interactions and why are they important? If you want to get answers on these questions (and many more), keep reading.
Knotty DNA
Try taking out your earphones from your pocket and, in all probability, you’ll find knots and entanglements between the ends. As it turns out, this knotting effect is not limited to macroscopic objects, but occurs on the nanoscale as well. A DNA molecule that carries the genetic information of a living organism is actually a long string-like polymer, so you can imagine that it would also get tangled up just like the cords of your earphones. In today’s paper, Calin Plesa and his colleagues at TU Delft are able to observe and measure these knots in DNA strands and uncover behaviour which has not been observed before.