For the most part of biology, it is form that follows function. Proteins are a perfect example of this — they are made of a sequence of amino acids (the protein building units), which are synthesized by the ribosome. Once synthesized, the long strings of amino acids fold up into a particular 3D shape or conformational state. Proteins take less than a thousandth of a second to attain their preferred conformational state (called “native state”) that — if nothing goes wrong — ends up being the same for a given sequence. This process is called protein folding. Explaining how a protein finds its folding preference out of all possible ways in such a short time is a longstanding problem in biology.
Knotty DNA
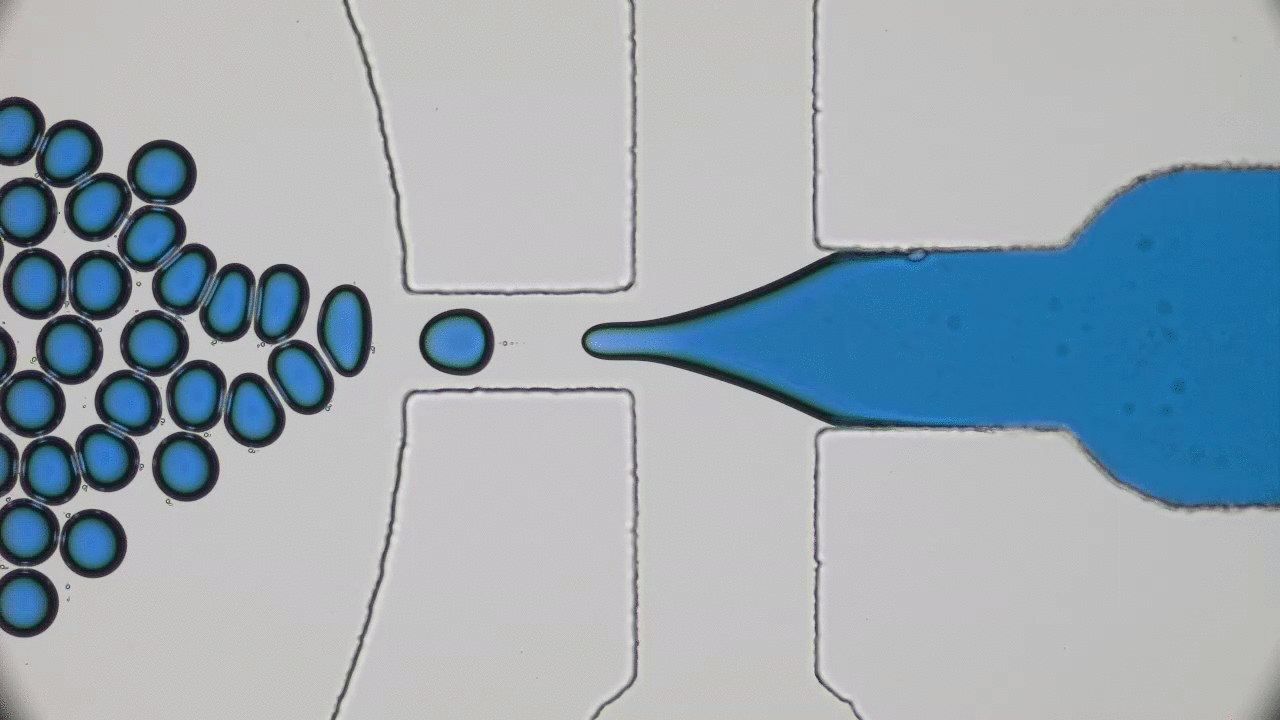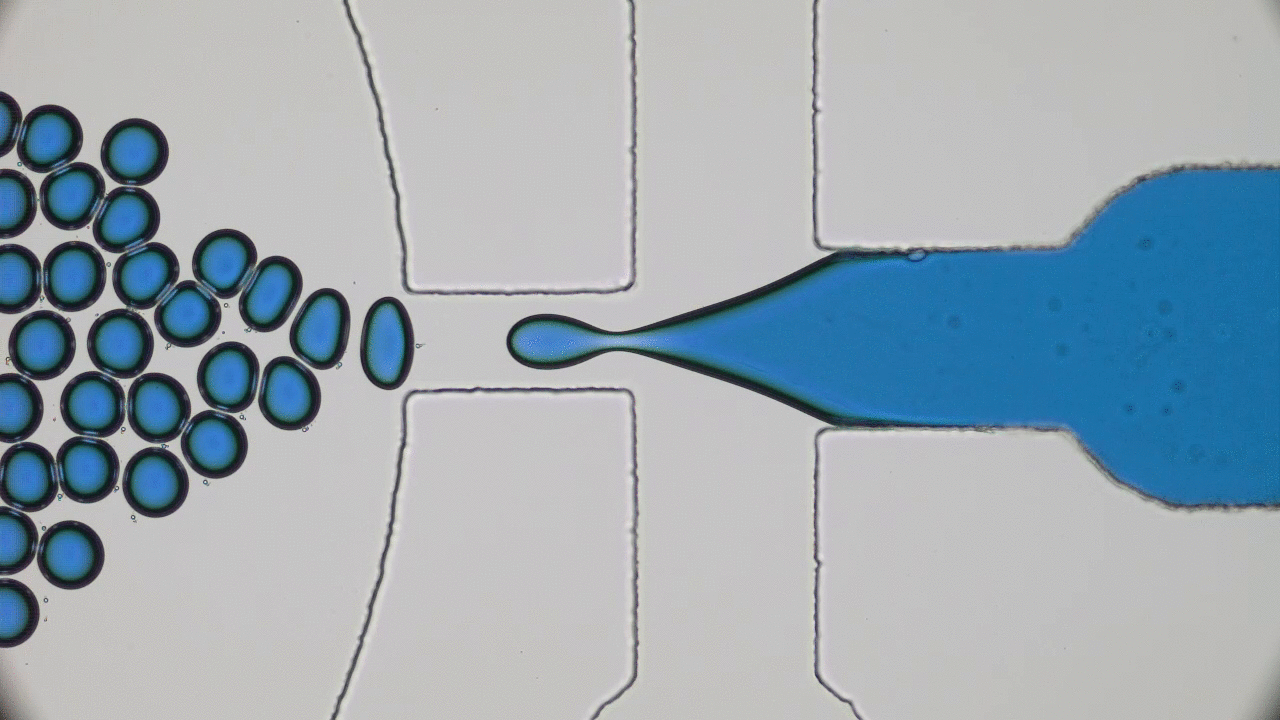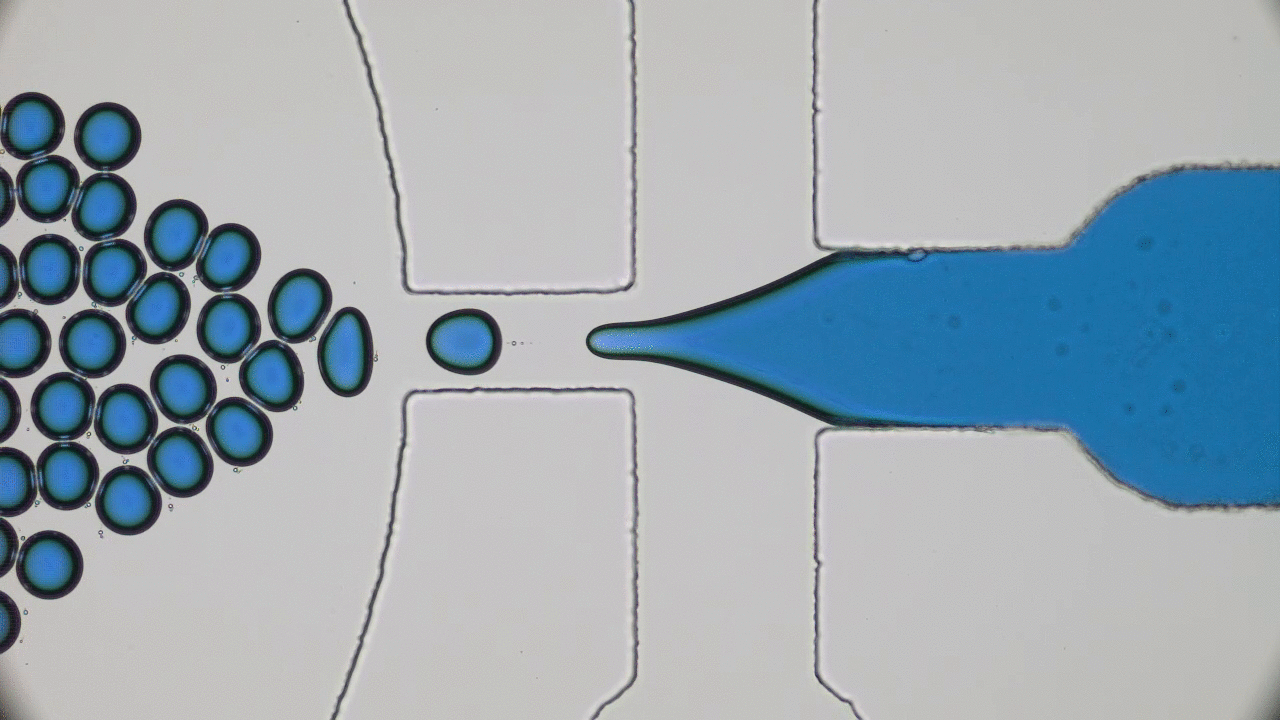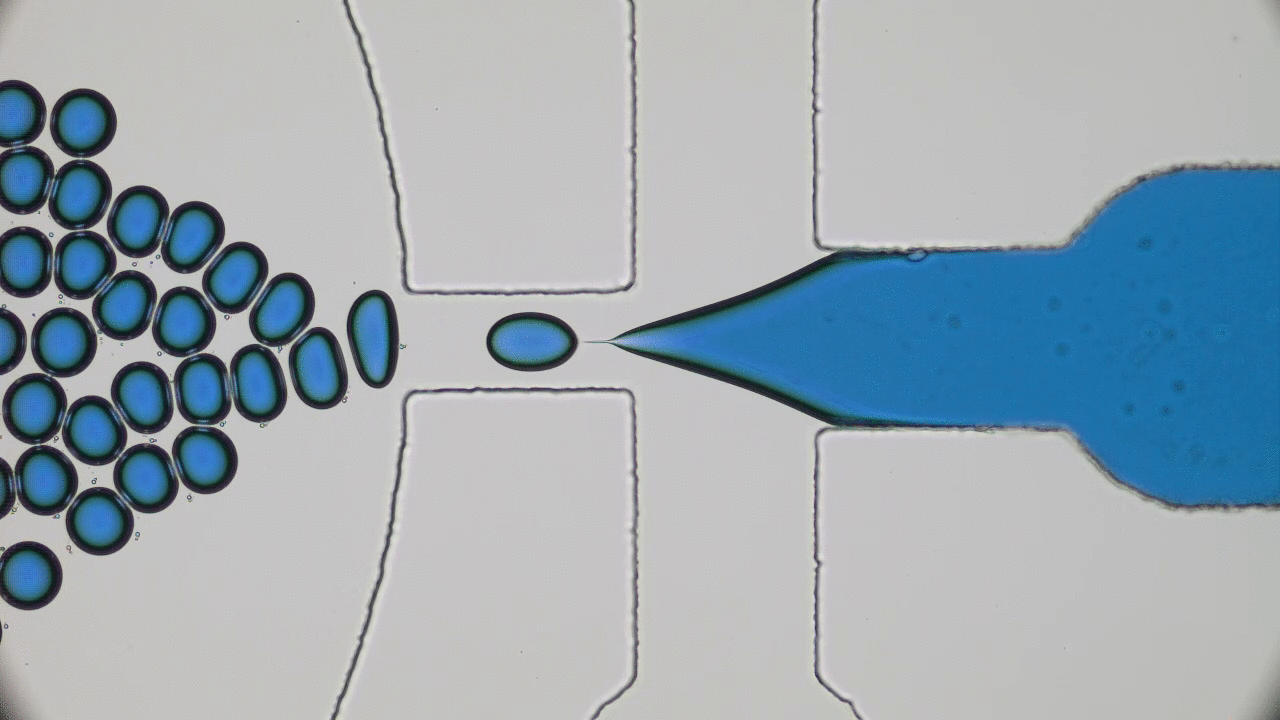
Try taking out your earphones from your pocket and, in all probability, you’ll find knots and entanglements between the ends. As it turns out, this knotting effect is not limited to macroscopic objects, but occurs on the nanoscale as well. A DNA molecule that carries the genetic information of a living organism is actually a long string-like polymer, so you can imagine that it would also get tangled up just like the cords of your earphones. In today’s paper, Calin Plesa and his colleagues at TU Delft are able to observe and measure these knots in DNA strands and uncover behaviour which has not been observed before.