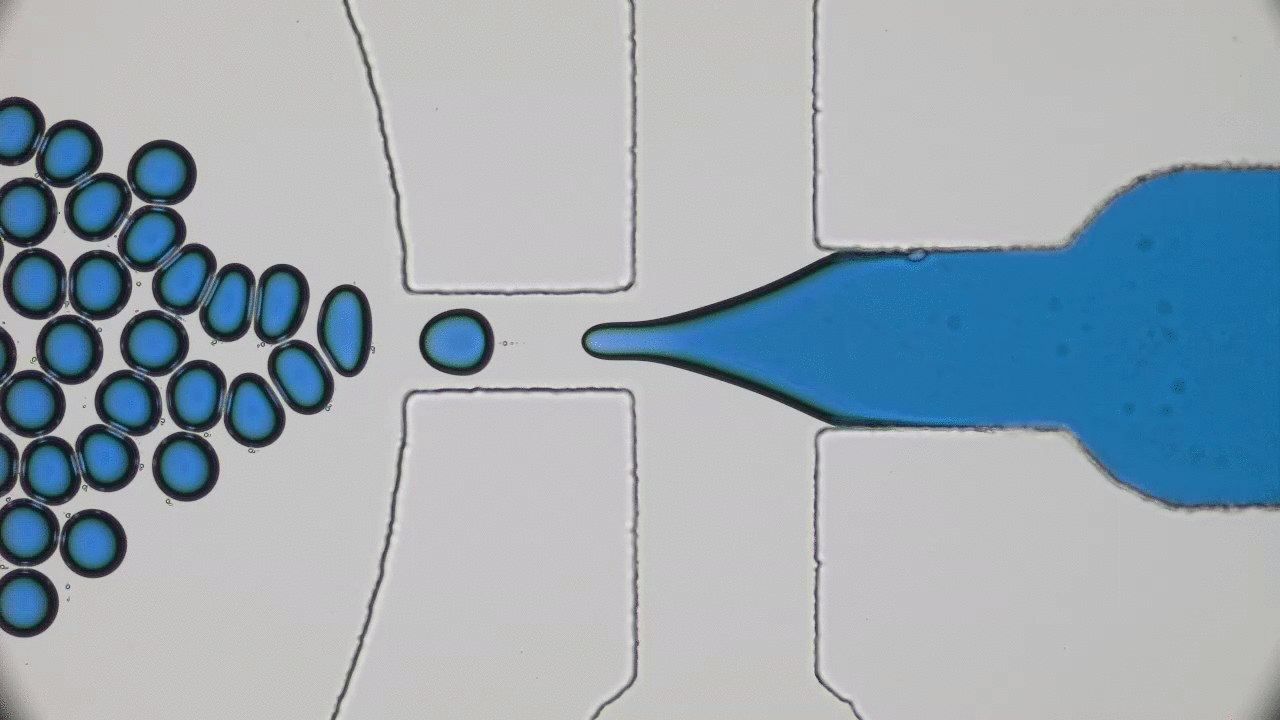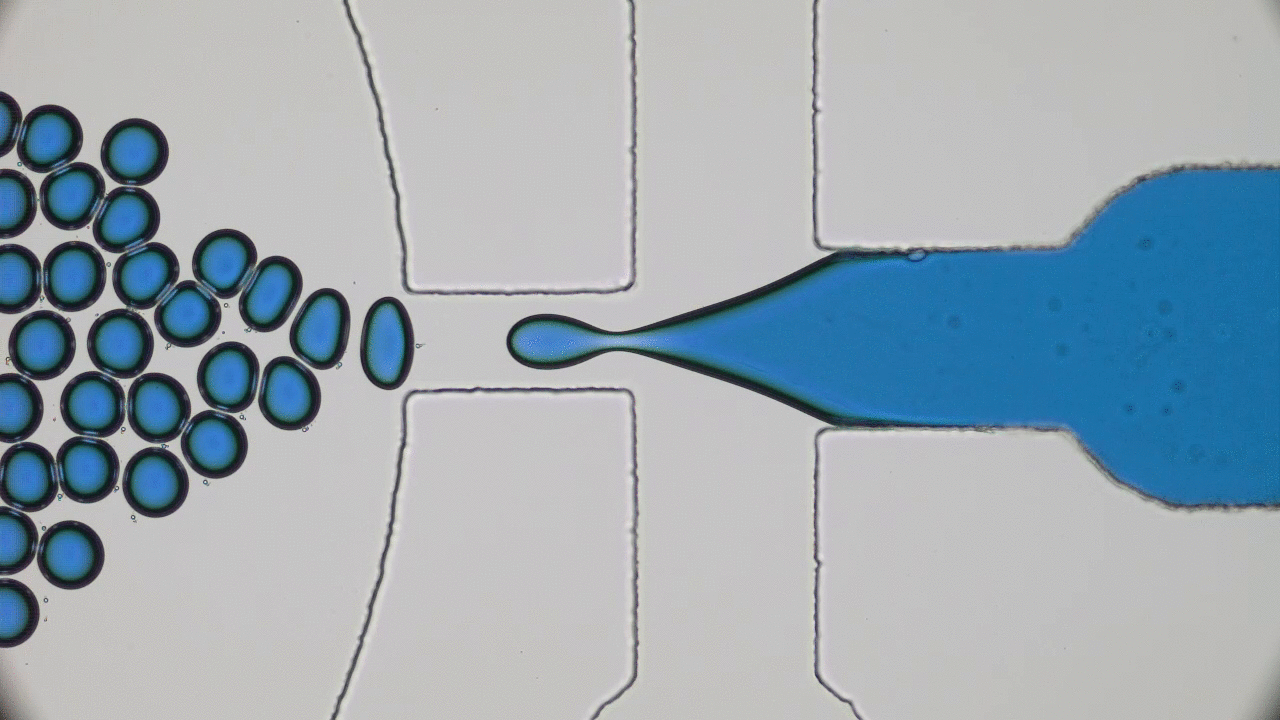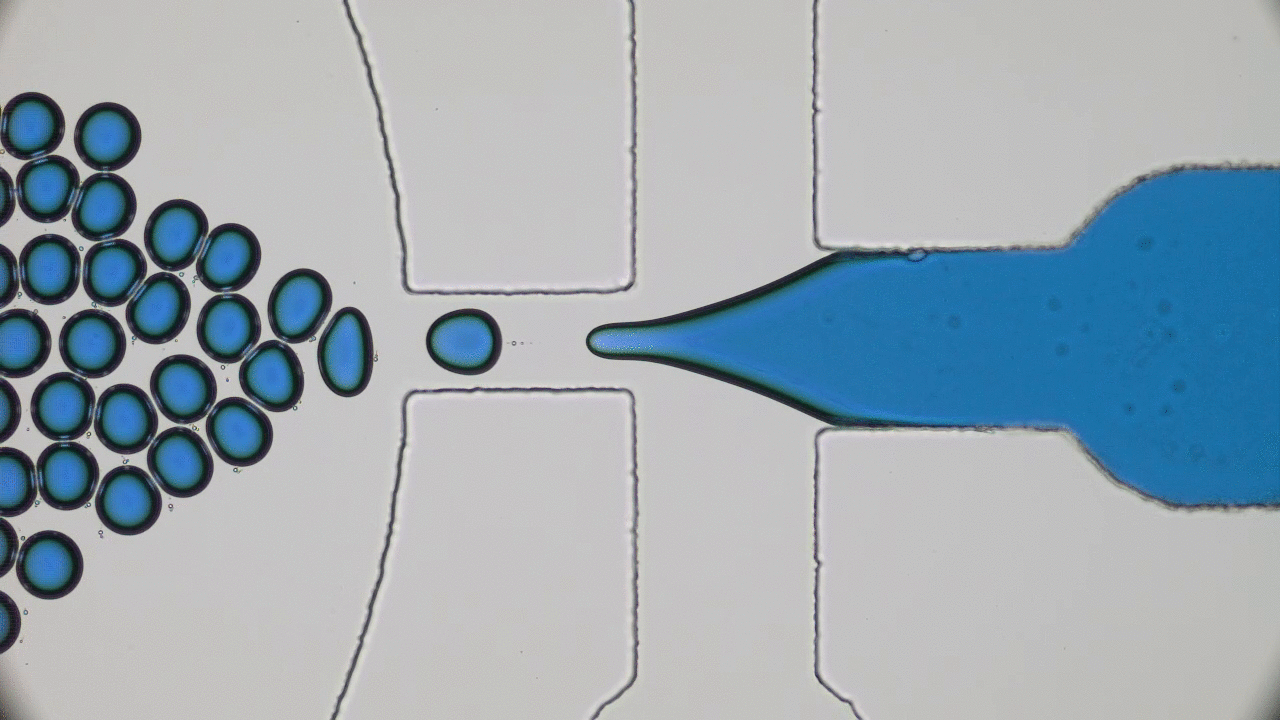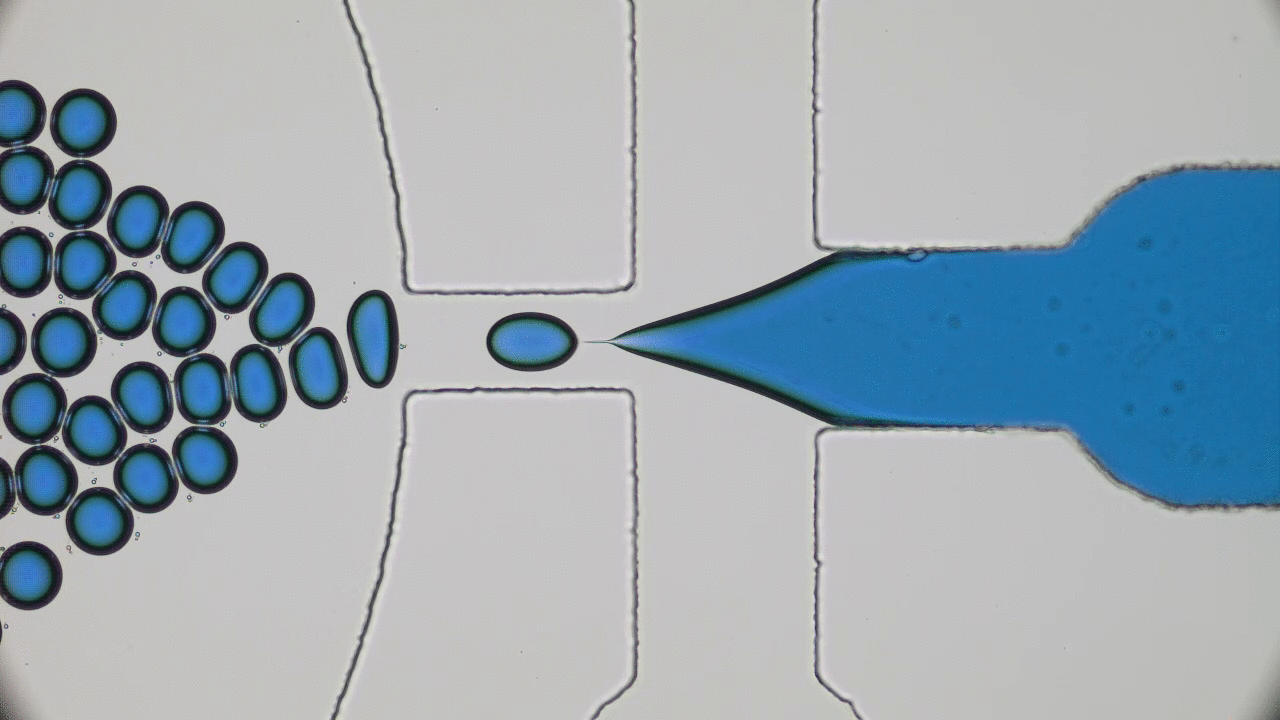
Understanding the origin of life is one of the most enduring and fundamental scientific challenges there is. Of all branches of science, physics is probably not the first place one would think to go to for enlightenment. Life seems too complicated and multi-layered to be captured by the simplistic frameworks of physics. Today’s paper tackles a small part of understanding the origin of life – the physics of self-replication.
Who needs polymer physics when you can get worms drunk instead?
If you speak to a soft matter physicist these days, within a few minutes the term “active matter” is bound to come up. A material is considered “active” when it burns energy to produce work, just like all sorts of molecular motors, proteins, and enzymes do inside your body. In this study, the scientists are focusing specifically on active polymers. These are long molecules which can burn energy to do physical work. Much of biological active matter is in the form of polymers (DNA or actin-myosin systems for example), and understanding them better would give direct insight into biophysics of all kinds. But polymers are microscopic objects with complex interactions, making them difficult to manipulate directly. To make matters worse, physicists have yet to fundamentally understand the behaviors of active materials, since they do not fit into our existing theories of so-called “passive” systems. In this study, Deblais and colleagues decided to entirely circumvent this problem by working with a much larger and easier-to-study system that behaves similarly to a polymer solution: a mixture of squirming worms in water.
Lifehack: How to pack two meters of chromatin into your cell’s nucleus, knot-free!
The entirety of our genetic information is encoded in our DNA. In our cells, it wraps together with proteins to form a flexible fiber about 2 metres long known as chromatin. Despite its length, each cell in our body keeps a copy of our chromatin in its nucleus, which is only about 10 microns across. For scale, if the nucleus was the size of a basketball, its chromatin would be about 90 miles long. How can it all fit in there? To make matters worse, the cell needs chromatin to be easily accessible for reading and copying, so it can’t be all tangled up. It’s not surprising then that scientists have been puzzled as to how this packing problem can be reliably solved in every cell. The solution is to pack the chromatin in a specific way, and research suggests that this may be in the form of a “fractal globule”.